Introduction
For those concerned about their cardiac health and researchers investigating in that field I'd like to present the ECG annotation library I developed during several years since my PhD studies. The last year I spent improving its quality of annotation during my post-doc research in Cambridge. It turned out that the work was not futile as I took the first place in Physionet Computers In Cardiology 2006 QT Annotation challenge. You may also have a look at the previous versions of the library at the physionet sources. The paper describing the algorithm available online Individually Adaptable Automatic QT Detector. Computers in Cardiology 2006. V. 33. PP. 337-341. You may have a look at the conference poster (Power Point 2007) I provided as a download at the top of this article.
I provided a console application to the library, so you can start with your ECG data and annotate it. You should have your data in Physionet ECG format, have a look at the site for a tool that convert text data files to it.
Background
You should have basics in cardiology, be familiar with physionet ECG data format and optionally with C++ programming and wavelet-analysis if you want to extend or modify the code or use wavelet-transforms classes I provided with my library. There is a nice tutorial by Robi Polikar available online on wavelet-transforms.
Using the Code
To annotate the ECG file, just run console application this way (I enclosed the physionet file n26c.dat with its header file n26c.hea from afpdb database for example):
>ecg.exe n26c.dat 1
This will annotate the first lead from the record.
>ecg.exe n26c.dat 2
To annotate the second lead, and so on.
Upon completion, the ECG annotation including P, T, QRS waves, ectopic beats and noise will be printed to stdout and saved to n26c.atr physionet compatible annotation file. Also HRV data will be saved in text format to n26c.hrv file and mean heart rate will be displayed at the stdout. To view the ECG record with the annotation you may browse physionet tools. I'll think of posting a simple GUI to view the record and the annotation.
As the records could be quite long in duration the output to stdout is quite slow so you may redirect it to the file for faster processing:
>ecg.exe n26c.dat 1 >output
The ECG annotation parameters used for specifying minimum, maximum ECG wave's intervals, durations, etc... are set to default values. I may post the modification with external set up for the console to fit them for particular ECG morphology, otherwise you may modify it yourself passing them to EcgAnnotation constructor.
The classes present in the library are:
Signal CWT : public Signal FWT : public Signal EcgDenoise : public FWT EcgAnnotation : public Signal
The Signal class provides reading ECG data in text, physionet and my custom file formats, saving it to disk, data normalization routines, denoising operations and some math functions. Have a look at the signal.h file.
The CWT class provides continuous wavelet transform. It is equipped with common wavelets (Mehihan Hat, Morlet, Gaussian derivatives). You can use it this way:
double SR;
double w0;
double* Data;
int size;
double* pSpec;
CWT cwt;
cwt.InitCWT(size, CWT::MHAT, w0, SR);
pSpec = cwt->CwtTrans(Data, 30.0);
pSpec = cwt->CwtTrans(Data, 45.56);
cwt.CloseCWT();
The FWT class provides 1D fast wavelet transform. You can use it in a similar way as CWT one.
wchar_t filter[] = L"path\\filters\\daub2.flt";
FWT fwt;
fwt.InitFWT(filter, Data, size);
fwt.FwtTrans(3);
pSpec = GetFwtSpectrum();
fwt.FwtSynth(3);
fwt.CloseFWT();
EcgDenoise is designed to denoise ECG signal with fast wavelet transform.
wchar_t path[] = L"fullpath\\filters";
EcgDenoise enoise;
enoise.InitDenoise(path, Data, size, SR);
enoise.LFDenoise();
enoise.HFDenoise();
enoise.LFHFDenoise();
enoise.CloseDenoise();
You may call denoising functions any number of times and in any order after initialization. Each time your Data will be filled with a denoised version of the signal.
EcgAnnotation is the class for complete ECG annotation. Here is the code from my console application on how to annotate the ECG data and save the results:
class Signal signal;
if (signal.ReadFile(argv[1])) {
int size = signal.GetLength();
double sr = signal.GetSR();
int h, m, s, ms;
int msec = int(((double)size / sr) * 1000.0);
signal.mSecToTime(msec, h, m, s, ms);
wprintf(L" leads: %d\n", signal.GetLeadsNum());
wprintf(L" sr: %.2lf Hz\n", sr);
wprintf(L" bits: %d\n", signal.GetBits());
wprintf(L" UmV: %d\n", signal.GetUmV());
wprintf(L" length: %02d:%02d:%02d.%03d\n\n", h, m, s, ms);
double* data = signal.GetData(leadNumber);
class EcgAnnotation ann;
wprintf(L" getting QRS complexes... ");
tic();
int** qrsAnn = ann.GetQRS(data, size, sr, L"filters");
if (qrsAnn) {
wprintf(L" %d beats.\n", ann.GetQrsNumber());
ann.GetEctopics(qrsAnn, ann.GetQrsNumber(), sr);
wprintf(L" getting P, T waves... ");
int annNum = 0;
int** ANN = ann.GetPTU(data, size, sr, L"filters",
qrsAnn, ann.GetQrsNumber());
if (ANN) {
annNum = ann.GetEcgAnnotationSize();
wprintf(L" done.\n");
toc();
wprintf(L"\n");
wcscpy(annName, argv[1]);
change_extension(annName, L".atr");
ann.SaveAnnotation(annName, ANN, annNum);
} else {
ANN = qrsAnn;
annNum = 2 * ann.GetQrsNumber();
wprintf(L" failed.\n");
toc();
wprintf(L"\n");
}
for (int i = 0; i < annNum; i++) {
int smpl = ANN[i][0];
int type = ANN[i][1];
msec = int(((double)smpl / sr) * 1000.0);
signal.mSecToTime(msec, h, m, s, ms);
wprintf(L"%10d %02d:%02d:%02d.%03d %s\n",
smpl, h, m, s, ms, anncodes[type]);
}
vector<double /> rrs;
vector<int> rrsPos;
wcscpy(hrvName, argv[1]);
change_extension(hrvName, L".hrv");
if (ann.GetRRseq(ANN, annNum, sr, &rrs, &rrsPos)) {
FILE *fp = _wfopen(hrvName, L"wt");
for (int i = 0; i < (int)rrs.size(); i++)
fwprintf(fp, L"%lf\n", rrs[i]);
fclose(fp);
wprintf(L"\n mean heart rate: %.2lf",
signal.Mean(&rrs[0], (int)rrs.size()));
}
} else {
wprintf(L"could not get QRS complexes.
make sure you have got \"filters\"
directory in the ecg application dir.");
exit(1);
}
} else {
wprintf(L"failed to read %s file", argv[1]);
exit(1);
}</int>
History
Update 1.0 - 28 Oct 2007
I modified console code and EcgAnnotation class after being asked by researchers using my code to provide more precise annotation for biphasic T waves and read annotation parameters from an external file. Now you may change the last setting in your parameters file biTwave from 0 to 1 to handle biphasic T waves. Changes concern EcgAnnotation::GetPTU() function there I added gaussian CWT filter for annotation of biphasic T waves.

You may run the console providing an additional optional third parameter as a file name with your particular annotation settings:
>ecg.exe patient1.dat 1 patient1params
>ecg.exe patient2.dat 1 patient2biTwave
>ecg.exe patient3.dat 2 patient3largeTwaves
Update 1.1 - 29 Oct 2007
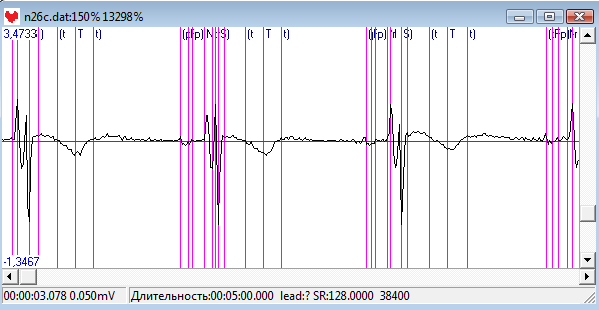
This one provides additional parameters loaded from an external file. With ampQRS set to 1 you can pre amplify QRS complex during the detection process. The qrsFreq denotes filtering frequency of CWT transform, default is 13Hz. The above mentioned changes allow to handle such abnormal ECGs as the one depicted below. As you can see the T wave is biphasic and peak shaped quite similar to QRS. With default params you cannot filter out such T wave and it spawns spurious detections of noise or another beat. The actual heart rate is 160 bpm, quite high. To deal with such abnormalities you have to increase qrsFreq filtering frequency to about 25Hz also with pre amplification of QRS complex ampQRS = 1. With those settings such T waves will be successfully filtered out and the QRS detection stage will be perfect.

Now the configuration file contains those fields:
minbpm 40
maxbpm 180
minQRS 0.04
maxQRS 0.2
qrsFreq 13
ampQRS 0
minUmV 0.2
minPQ 0.07
maxPQ 0.20
minQT 0.22
maxQT 0.45
pFreq 9.0
tFreq 3.0
biTwave 0
