Introduction
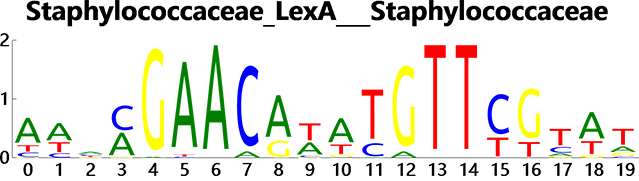
Picture1. Sequence logo draw for LexA in Staphylococcaceae from RegPrecise database: http://regprecise.lbl.gov/RegPrecise/regulog.jsp?regulog_id=680
In bioinformatics, a sequence logo is a graphical representation of the sequence conservation of nucleotides (in a strand of DNA/RNA) or amino acids (in protein sequences). A sequence logo is created from a collection of aligned sequences and depicts the consensus sequence and diversity of the sequences. Sequence logos are frequently used to depict sequence characteristics such as protein-binding sites in DNA or functional units in proteins.(https://en.wikipedia.org/wiki/Sequence_logo)
In this article I want to introduce how to creates this sequence logo diagram using VisualBasic language.
Background
There are many tools can creates the motifs model for the sequence logo drawing. A sequence logo model can be simply creates from the multiple sequence alignment by using MEGA or Clustal software if the data requirements is not so strict; but in more often situation, the motif should creates from the PWM, and the meme suite software is doing the best in motifs PWM discovery and creation.

Picture2. Drawing a sequence logo in GCModeller
Some concepts in the motifs
1. Fasta sequence
The most common used database format to stored the biological sequence in text files, one fasta sequence is start with the title line, which its first character must be >
And then starts from the second line, is the biological sequence data, here is a example of a simple nucleotide sequence:
>XC_0012:21_Sigma54
-------TGCTGCTGTT--GCTGC---
>XC_0013:35_Sigma54
-------AGGCACAGGAAGTGC-----
A parser for the fasta sequence database is already implemented in GCModeller for .NET language, which is available at namespace: LANS.SystemsBiology.SequenceModel.FASTA.FastaFile
Use this parser just simply enough:
Imports LANS.SystemsBiology.SequenceModel
Dim fasta As New FASTA.FastaFile(path)
2. Alphabet Relative Frequency
The alphabets between the protein and DNA is different, for DNA sequence just ATGC 4 alphabets, and for protein sequence, there are 20 characters:
Public ReadOnly Property NT As IReadOnlyCollection(Of Char) = {"A"c, "T"c, "G"c, "C"c}
Public ReadOnly Property AA As IReadOnlyCollection(Of Char) = {
"A"c, "R"c, "N"c, "D"c, "C"c, "E"c, "Q"c, "G"c,
"H"c, "I"c, "L"c, "K"c, "M"c, "F"c, "P"c, "S"c,
"T"c, "W"c, "Y"c, "V"c
}

Picture3. Statistics of the alphabet relative frequency
For calculate the alphabet relative frequency, just simply statics the frequency of each alphabet that occurs in each column. And here is the simply version of statics the alphabet frequency:
- counts for the alphabets its number of occurrences in a column;
- then using the occurrences number Divide the total number of the sequence
Imports Microsoft.VisualBasic.Linq
Imports LANS.SystemsBiology.SequenceModel.FASTA
<ExportAPI("NT.Frequency")>
Public Function Frequency(source As IEnumerable(Of FastaToken)) As PatternModel
Dim len As Integer = source.First.Length
Dim n As Integer = source.Count
Dim alphabets As Char() =
If(source.First.IsProtSource,
Polypeptides.ToChar.Values.ToArray, New Char() {"A"c, "T"c, "G"c, "C"c})
Dim fasta As New FastaFile(source.ToArray(Function(x) x.ToUpper))
Dim LQuery = (From pos As Integer
In len.Sequence.AsParallel
Select pos,
row = (From c As Char
In alphabets
Select c,
f = __frequency(fasta, pos, c, n)).ToArray
Order By pos Ascending).ToArray
Dim Model As IEnumerable(Of SimpleSite) =
From x
In LQuery.SeqIterator
Let freq As Dictionary(Of Char, Double) =
x.obj.row.ToDictionary(Function(o0) o0.c, Function(o0) o0.f)
Select New SimpleSite(freq, x.Pos)
Return New PatternModel(Model)
End Function
Private Function __frequency(Fasta As IEnumerable(Of FastaToken),
p As Integer,
C As Char,
numOfFasta As Integer) As Double
Dim LQuery As Integer = (From nt As FastaToken
In Fasta
Let chr As Char = nt.SequenceData(p)
Where C = chr
Select 1).Sum
Dim f As Double = LQuery / numOfFasta
Return f
End Function
This alphabet frequency statics function code can be found at namespace: LANS.SystemsBiology.SequenceModel.Patterns.PatternsAPI
Using the code
As describe in the picture 2, for drawing a sequence logo in VisualBasic, we should creates a drawing model for the visualization. And for drawing a sequence logo, there are 3 type of information that we needs for the diagram:
- alphabets of the residues
- height of each alphabet in a column
- the columns of the motif that this sequence logo diagram will represents
Alphabets rendering
First of all, let see how to rendering the residue alphabets:
Due to the reason of there are two types of the sequence that can be store in the fasta sequence database, so that we needs two color schema for the sequence logo visualization corresponding to the DNA sequence and protein sequence. Here are the colors definitions
Color schema for the nucleotide sequence:
Public ReadOnly Property NucleotideSchema As Dictionary(Of Char, Image)
Get
Return New Dictionary(Of Char, Image) From {
{"A"c, ColorSchema.__getTexture(Color.Green, "A")},
{"T"c, ColorSchema.__getTexture(Color.Red, "T")},
{"G"c, ColorSchema.__getTexture(Color.Yellow, "G")},
{"C"c, ColorSchema.__getTexture(Color.Blue, "C")}
}
End Get
End Property
Color schema for the protein residues alphabets:
Public ReadOnly Property ProteinSchema As Dictionary(Of Char, Image)
Get
Return New Dictionary(Of Char, Image) From {
{"A"c, ColorSchema.__getTexture(Color.CadetBlue, "A")},
{"R"c, ColorSchema.__getTexture(Color.Black, "R")},
{"N"c, ColorSchema.__getTexture(Color.Chocolate, "N")},
{"D"c, ColorSchema.__getTexture(Color.Coral, "D")},
{"C"c, ColorSchema.__getTexture(Color.Chartreuse, "C")},
{"E"c, ColorSchema.__getTexture(Color.Cyan, "E")},
{"Q"c, ColorSchema.__getTexture(Color.LawnGreen, "Q")},
{"G"c, ColorSchema.__getTexture(Color.DarkMagenta, "G")},
{"H"c, ColorSchema.__getTexture(Color.Gold, "H")},
{"I"c, ColorSchema.__getTexture(Color.HotPink, "I")},
{"L"c, ColorSchema.__getTexture(Color.LightSlateGray, "L")},
{"K"c, ColorSchema.__getTexture(Color.Yellow, "K")},
{"M"c, ColorSchema.__getTexture(Color.Teal, "M")},
{"F"c, ColorSchema.__getTexture(Color.SaddleBrown, "F")},
{"P"c, ColorSchema.__getTexture(Color.Red, "P")},
{"S"c, ColorSchema.__getTexture(Color.RoyalBlue, "S")},
{"T"c, ColorSchema.__getTexture(Color.Tomato, "T")},
{"W"c, ColorSchema.__getTexture(Color.MediumSeaGreen, "W")},
{"Y"c, ColorSchema.__getTexture(Color.SkyBlue, "Y")},
{"V"c, ColorSchema.__getTexture(Color.Maroon, "V")}
}
End Get
End Property
Due to the reason of it is hardly to controls the string drawing style directly using DrawString method on the sequence logo diagram, and the method DrawImage of gdi+ can directly controls the image drawing location and size through x, y, width and heigh these function parameters, so that I creates the image cache for each alphabet which is required in the logo alphabets at first. and then these alphabet's image cache can makes the later logo drawing program more easily. Here is the code of transforming the alphabet string into a image cache:
Private Function __getTexture(color As Color, alphabet As String) As Image
Dim bitmap As New Bitmap(680, 680)
Dim font As New Font(FontFace.Ubuntu, 650)
Dim br As New SolidBrush(color:=color)
Using gdi As Graphics = Graphics.FromImage(bitmap)
Dim size As SizeF = gdi.MeasureString(alphabet, font:=font)
Dim w As Integer = (bitmap.Width - size.Width) / 2
Dim h As Integer = (bitmap.Height - size.Height) * 0.45
Dim pos As New Point(w, h)
gdi.CompositingQuality = Drawing2D.CompositingQuality.HighQuality
gdi.CompositingMode = Drawing2D.CompositingMode.SourceOver
Call gdi.DrawString(alphabet, font, br, point:=pos)
End Using
Return bitmap
End Function
These code can be found at module:
LANS.SystemsBiology.AnalysisTools.SequenceTools.SequencePatterns.SequenceLogo.ColorSchema
Calculation of the Bits of residue sites
One of the important thing is measure the alphabet height that draw on the sequence logo. The height of the entire stack of residues is the information measured in bits. And this value is relative to the frequency of the alphabets and the number of the sequence from the fasta source, and reference from the Wikipedia, a formula was given to calculate this information bits:

Where the value of base is relative to the residue type, base is 4 for DNA sequence and 20 for protein sequence. For instance, here is the code example:
Dim base As Integer = If(fasta.First.IsProtSource, 20, 4)
E its value is relative to the base too, and the it also relatives to the number of the fasta sequence in the alignment source:

Where variable n is the number of the fasta sequence in the alignment, which can be achieve from a property in FastaFile class:
Dim n As Integer = FastaFile.NumberOfFasta
Dim base As Integer = If(fa.First.IsProtSource, 20, 4)
Dim E As Double = (1 / Math.Log(2)) * ((base - 1) / (2 * n))
And the uncertain information for the bits of each residue site i that can be calculate from:

Where i is the column in the motif matrix, and f(a) is the alphabet relative frequency that we've creates previously. and a is the alphabet, where the relative frequency function its parameter a belongs to "ATGC" if the sequence is DNA, otherwise is the 20 amino acid residue alphabet for protein sequence. Codes for calculate this uncertain information Hi is show below:
Imports Microsoft.VisualBasic.Linq
Dim H As Double() = f.Residues.ToArray(Function(x) x.Alphabets.__hi)
<Extension>
Private Function __hi(f As Dictionary(Of Char, Double)) As Double
Dim h As Double = f.Values.Sum(Function(n) If(n = 0R, 0, n * Math.Log(n, 2)))
h = 0 - h
Return h
End Function
Then when we calculated the bits information for each residue, with the previous statics position relative alphabet frequency matrix, then we have enough information to descript a sequence motif as sequence logo.
Here is the whole code for creates the frequency motif model, which is available at namespace: LANS.SystemsBiology.AnalysisTools.SequenceTools.SequencePatterns.Motif.PWM
Imports System.Runtime.CompilerServices
Imports LANS.SystemsBiology.AnalysisTools.SequenceTools.SequencePatterns.SequenceLogo
Imports LANS.SystemsBiology.SequenceModel.FASTA
Imports LANS.SystemsBiology.SequenceModel.Patterns
Imports Microsoft.VisualBasic.Language
Imports Microsoft.VisualBasic.Linq
Public Function FromMla(fa As FastaFile) As MotifPWM
Dim f As PatternModel = PatternsAPI.Frequency(fa)
Dim n As Integer = fa.NumberOfFasta
Dim base As Integer = If(fa.First.IsProtSource, 20, 4)
Dim E As Double = (1 / Math.Log(2)) * ((base - 1) / (2 * n))
Dim H As Double() = f.Residues.ToArray(Function(x) x.Alphabets.__hi)
Dim PWM As ResidueSite() =
LinqAPI.Exec(Of SimpleSite, ResidueSite) _
(f.Residues) <= Function(x, i) __residue(x.Alphabets, H(i), E, base, i)
If base = 20 Then
Return MotifPWM.AA_PWM(PWM)
Else
Return MotifPWM.NT_PWM(PWM)
End If
End Function
Private Function __residue(f As Dictionary(Of Char, Double), h As Double, en As Double, n As Integer, i As Integer) As ResidueSite
Dim R As Double = Math.Log(n, 2) - (h + en)
Dim alphabets As Double()
If n = 4 Then
alphabets = {f("A"c), f("T"c), f("G"c), f("C"c)}
Else
alphabets = LinqAPI.Exec(Of Double) <= From c As Char In ColorSchema.AA Select f(c)
End If
Return New ResidueSite With {
.Bits = R,
.PWM = alphabets,
.Site = i
}
End Function
<Extension>
Private Function __hi(f As Dictionary(Of Char, Double)) As Double
Dim h As Double = f.Values.Sum(Function(n) If(n = 0R, 0, n * Math.Log(n, 2)))
h = 0 - h
Return h
End Function
Build a Drawing Model
As the article described previously, for drawing a sequence logo, we should know what alphabets and their relative frequency in each column position. For measuring the height of each residue site, we should calculate the bits information based on the alphabet relative frequency and the number of th aligned sequence. So that after the data model calculation, all of the information that required for the logo drawing is achieved. For the further unified operation of sequence logo drawing from both multiple sequence alignment result and PWM motif model, so that we should transform the frequency model or PWM into a unify DrawingModel. And here is the DrawingModel class type definitions:
DrawingModel for represents the whole motif's data modelResidue for each column site in the motif sites- And the relative frequency of each column is consist of the
Alphabet class
Public Class DrawingModel : Inherits ClassObject
Public Property Residues As Residue()
Public Property En As Double
Public Property ModelsId As String
Public Overrides Function ToString() As String
Return ModelsId & " --> " & Me.GetJson
End Function
End Class
Public Class Residue : Implements IAddressHandle
Public Property Alphabets As Alphabet()
Public Property Bits As Double
Public Property Address As Integer Implements IAddressHandle.Address
End Class
Public Class Alphabet : Implements IComparable
Public Property Alphabet As Char
Public Property RelativeFrequency As Double
End Class
These class type definition is available at namespace: LANS.SystemsBiology.AnalysisTools.SequenceTools.SequencePatterns.SequenceLogo
That is all we needs to represent a sequence logo in the data model. And this function provides the interface for convert the relative frequency model into the DrawingModel andalso the motif sequence logo drawing at the end of the function:
<ExportAPI("Drawing.Frequency")>
Public Function DrawFrequency(Fasta As FastaFile, Optional title As String = "") As Image
Dim PWM As MotifPWM = Motif.PWM.FromMla(Fasta)
Dim Model As DrawingModel = New DrawingModel
#If DEBUG Then
Dim m As String = New String(PWM.PWM.ToArray(Function(r) r.AsChar))
Call VBDebugger.WriteLine(m, ConsoleColor.Magenta)
#End If
If String.IsNullOrEmpty(title) Then
If Not String.IsNullOrEmpty(Fasta.FileName) Then
Model.ModelsId = Fasta.FileName.BaseName
Else
Model.ModelsId = New String(PWM.PWM.ToArray(Function(r) r.AsChar))
End If
Else
Model.ModelsId = title
End If
Model.Residues =
LinqAPI.Exec(Of ResidueSite, Residue)(PWM.PWM) <=
Function(rsd As ResidueSite) New Residue With {
.Bits = rsd.Bits,
.Address = rsd.Site,
.Alphabets = LinqAPI.Exec(Of Alphabet) <= From x As SeqValue(Of Double)
In rsd.PWM.SeqIterator
Select New Alphabet With {
.Alphabet = PWM.Alphabets(x.Pos),
.RelativeFrequency = x.obj
}
}
Return InvokeDrawing(Model, True)
End Function

Here are two picture to shows the relationship between the components of the DrawingModel:

Picture4. The relationship between the residue and its alphabets.
The sequence logo was consists of a sequence of residue sites, where each residue site is consists of all of the alphabets of the sequence, but the relative frequency of each alphabet in a residue is usually not equals to each other

Picture5. The relationship between the residue and the DrawingModel class
One residue is consists of all of the alphabets, and then the DrawingModel is a sequence of these residue, each residue site its stack height is measure by the Bits information content, and the bits information content is related to the alphabet relative frequency in each residue site.
Sequence logo drawer function
As we mentioned above, the alphabet height is related to the Bits information content on one residue site, and the actual drawing height can be calculate from the product between the alphabet its relative frequency and the residue site:

and code for this alphabet height calculation:
YHeight = (n * DrawingDevice.Height) * (If(residue.Bits > MaxBits, MaxBits, residue.Bits) / MaxBits)
Dim H As Single = Alphabet.RelativeFrequency * YHeight

Picture6. Using the image drawing is more easily controls the location and size of the alphabet than draw string directly.
Y -= H
gdi.Gr_Device.DrawImage(
colorSchema(Alphabet.Alphabet),
CSng(X), CSng(Y),
CSng(DrawingDevice.WordSize), H)
The full code of the sequence logo drawing function can be found at namespace:
LANS.SystemsBiology.AnalysisTools.SequenceTools.SequencePatterns.SequenceLogo.DrawingDevice
Using the Example
The code in this article is already implemented as a CLI utility command in a sequence tools. Which the tools can be download from the github release. You can try to draw a sequence logo with the example test data, which is available at location https://github.com/SMRUCC/Sequence-Patterns-Toolkit/blob/master/data/Xanthomonadales_MetR___Xanthomonadales.fasta, and this motifs sequence data is download from RegPrecise database: http://regprecise.lbl.gov/RegPrecise/regulog.jsp?regulog_id=5789

Open the CMD terminal, and then using command seqtools ? /logo to get the details information about how to use this command
Here is a example commandline usage of drawing the example test data:
seqtools /logo /in ./data/Staphylococcaceae_LexA___Staphylococcaceae.fasta

