Introduction
Gauss-Newton algorithm is a mathematical model to solve non-linear functions. A simple non-linear function is given below:
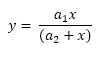
where a1 and a2 are the unknown parameters of this function. To find these two parameters, the values of y are measured on different values of x; y can be the rate of a chemical reaction and x is the concentration of a chemical affecting the rate. The results may look like this:
x | 0.038 | 0.194 | 0.425 | 0.626 | 1.253 | 2.500 | 3.740 |
y | 0.050 | 0.127 | 0.094 | 0.2122 | 0.2729 | 0.2665 | 0.3317 |
Gauss-Newton algorithm is used to estimate the values of a1 and a2 using the observed data above.
This sample function and the data are taken from https://en.wikipedia.org/wiki/Gauss-Newton_algorithm.
Background
Gauss-Newton algorithm in a few steps:
- Create an array of unknown parameters in the function:
b = (b1, b2,...,bn)
- Initialise the parameters and for each data point in matrix x, calculate the predicted values (y').
- Calculate the residuals:
ri = y'i - yi
- Find the partial differential of residuals with respect to the parameters and generate Jacobian matrix.
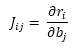
- Following an iterative process, calculate the new values for the parameters using the following equation:
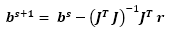
s is the iteration number, J is the Jacobian matrix, JT is the transpose of J.
For more information, please visit https://en.wikipedia.org/wiki/Gauss-Newton_algorithm.
Using the Code
In this implementation, I will be using some matrix operations that have already been implemented in another article. For more information, please take a look at this article.
- Given matrix x and y, the optimization starts with an initial guess of b matrix. By default, a value of
1.0 is given to all parameters in non-linear function. - Calculate the residuals:
public double[][] calculateResiduals(double[][] x, double[] y, double[] b) {
double[][] res = new double[y.length][1];
for (int i = 0; i < res.length; i++) {
res[i][0] = findY(x[i][0], b) - y[i];
}
return res;
}
For each data point in x matrix, function findY() is called to calculate the predicted value of y. In the code, you will find this function as abstract:
public abstract double findY(double x, double[] b);
The user needs to implement this function before using the optimization. For example, for the function in the introduction, the implementation will be as follows:
GaussNewton gaussNewton = new GaussNewton() {
@Override
public double findY(double x, double[] b) {
return (x * b[0]) / (b[1] + x);
}
};
- Calculate the Jacobian matrix which is the partial derivatives of the residuals with respect to parameters in the function:
public double[][] jacob(double[] b, double[][] x, int numberOfObservations) {
int numberOfVariables = b.length;
double[][] jc = new double[numberOfObservations][numberOfVariables];
for (int i = 0; i < numberOfObservations; i++) {
for (int j = 0; j < numberOfVariables; j++) {
jc[i][j] = derivative(x[i][0], b, j);
}
}
return jc;
}
Function derivative() is called to calculate the partial derivative:
public double derivative(double x, double[] b, int bIndex) {
double[] bCopy = b.clone();
bCopy[bIndex] += alpha;
double y1 = findY(x, bCopy);
bCopy = b.clone();
bCopy[bIndex] -= alpha;
double y2 = findY(x, bCopy);
return (y1 - y2) / (2 * alpha);
}
This function gives a good approximate of the partial derivative; i.e., the change in y after a small change in the variable.
- Perform (J JT)-1 JT r
operations:
public double[][] transjacob(double[][] JArray, double[][] res) throws NoSquareException {
Matrix r = new Matrix(res);
Matrix J = new Matrix(JArray);
Matrix JT = MatrixMathematics.transpose(J);
Matrix JTJ = MatrixMathematics.multiply(JT, J);
Matrix JTJ_1 = MatrixMathematics.inverse(JTJ);
Matrix JTJ_1JT = MatrixMathematics.multiply(JTJ_1, JT);
Matrix JTJ_1JTr = MatrixMathematics.multiply(JTJ_1JT, r);
return JTJ_1JTr.getValues();
}
- Using the results of step 4, the new values of the parameters are calculated:
IntStream.range(0, values.length).forEach(j -> b2[j] = b2[j] - gamma * values[j][0]);
b2 is the new b matrix. gamma is a fraction of the values coming from Jacobian. If the initial values for b are far from optimum, there is a possibility of convergence problem. Applying this simple fraction seems to solve this issue. The downside of applying this fraction is the number of iterations which will increase.
- Having the new b matrix, repeat steps 2-5 in the next iteration. All optimisation steps are given in the function below:
public double[] optimise(double[][] x, double[] y, double[] b) throws NoSquareException {
int maxIteration = 1000;
double oldError = 100;
double precision = 1e-6;
double[] b2 = b.clone();
double gamma = .01;
for (int i = 0; i < maxIteration; i++) {
double[][] res = calculateResiduals(x, y, b2);
double error = calculateError(res);
System.out.println("Iteration : " + i + ", Error-diff: " +
(Math.abs(oldError - error)) + ", b = "+ Arrays.toString(b2));
if (Math.abs(oldError - error) <= precision) {
break;
}
oldError = error;
double[][] jacobs = jacob(b2, x, y.length);
double[][] values = transjacob(jacobs, res);
IntStream.range(0, values.length).forEach(j -> b2[j] = b2[j] - gamma * values[j][0]);
}
return b2;
}
Example
Function
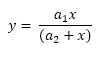
Observations
x | 0.038 | 0.194 | 0.425 | 0.626 | 1.253 | 2.500 | 3.740 |
y | 0.050 | 0.127 | 0.094 | 0.2122 | 0.2729 | 0.2665 | 0.3317 |
Target
Find a1 and a2 using the above data and Gauss-Newton algorithm:
@Test
public void optimiseWithInitialValueOf1() throws NoSquareException {
double[][] x = new double[7][1];
x[0][0] = 0.038;
x[1][0] = 0.194;
x[2][0] = 0.425;
x[3][0] = 0.626;
x[4][0] = 1.253;
x[5][0] = 2.500;
x[6][0] = 3.740;
double[] y = new double[] { 0.050, 0.127, 0.094, 0.2122, 0.2729, 0.2665, 0.3317 };
GaussNewton gaussNewton = new GaussNewton() {
@Override
public double findY(double x, double[] b) {
return (x * b[0]) / (b[1] + x);
}
};
double[] b = gaussNewton.optimise(x, y, 2);
Assert.assertArrayEquals(new double[]{0.36, 0.56}, b, 0.01);
}
In the above test, the initial values for the parameters are the default value (1.0); however, with starting values of 100, we will get the same values; see the attached codes. I have also tested the algorithm on a large set of randomly generated data and it works with good time and memory efficiency; see the test codes attached.
History
This is the first version of this article.
