In the previous post, we have seen how to calculate some of the basic parameters of descriptive statistics, as well as how to normalize data by calculating mean and standard deviation. In this blog post, we are going to implement data normalization as regular neural network layer, which can simplify the training process and data preparation.
What is Data Normalization?
Simply said, data normalization is a set of tasks which transform values of any feature in a data set into predefined number range. Usually, this range is [-1,1] , [0,1] or some other specific ranges. Data normalization plays a very important role in ML, since it can dramatically improve the training process, and simplify settings of network parameters.
There are two main types of data normalization:
- MinMax normalization – which transforms all values into range of
[0,1], - Gauss Normalization or Z score normalization, which transforms the value in such a way that the average value is zero, and std is
1.
Beside those types, there are plenty of other methods which can be used. Usually, those two are used when the size of the data set is known, otherwise we should use some of the other methods, like log scaling, dividing every value with some constant, etc. But why data needs to be normalized? This is the essential question in ML, and the simplest answer is to provide the equal influence to all features to change the output label. More about data normalization and scaling can be found at this link.
In this blog post, we are going to implement CNTK neural network which contains a “Normalization layer” between input and first hidden layer. The schematic picture of the network looks like the following image:
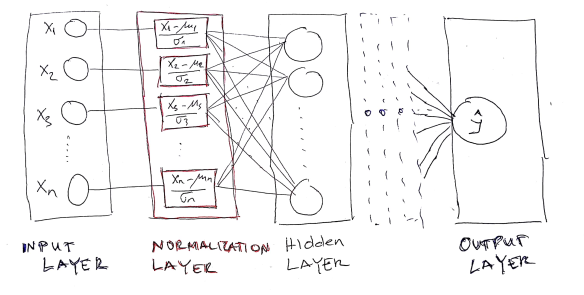
As can be observed, the Normalization layer is placed between input and first hidden layer. Also, the Normalization layer contains the same neurons as input layer and produced the output with the same dimension as the input layer.
In order to implement Normalization layer, the following requirements must be met:
- calculate average
 and standard deviation
and standard deviation  in training data set as well as find maximum and minimum value of each feature.
in training data set as well as find maximum and minimum value of each feature. - this must be done prior to neural network model creation, since we need those values in the normalization layer.
- within network model creation, the normalization layer should be defined after input layer is defined.
Calculation of Mean and Standard Deviation for Training Data Set
Before network creation, we should prepare mean and standard deviation parameters which will be used in the Normalization layer as constants. Hopefully, the CNTK has the static method in the Minibatch source class for this purpose “MinibatchSource.ComputeInputPerDimMeansAndInvStdDevs”. The method takes the whole training data set defined in the minibatch and calculates the parameters.
var d = new DictionaryNDArrayView, NDArrayView>>();
using (var mbs = MinibatchSource.TextFormatMinibatchSource(
trainingDataPath , streamConfig, MinibatchSource.FullDataSweep,false))
{
d.Add(mbs.StreamInfo("feature"), new Tuple(null, null));
MinibatchSource.ComputeInputPerDimMeansAndInvStdDevs(mbs, d, device);
}
Now that we have average and std values for each feature, we can create network with normalization layer. In this example, we define simple feed forward NN with 1 input, 1 normalization, 1 hidden and 1 output layer.
private static Function createFFModelWithNormalizationLayer
(Variable feature, int hiddenDim,int outputDim, Tuple<NDArrayView, NDArrayView> avgStdConstants,
DeviceDescriptor device)
{
var glorotInit = CNTKLib.GlorotUniformInitializer(
CNTKLib.DefaultParamInitScale,
CNTKLib.SentinelValueForInferParamInitRank,
CNTKLib.SentinelValueForInferParamInitRank, 1);
var inputLayer = feature;
var mean = new Constant(avgStdConstants.Item1, "mean");
var std = new Constant(avgStdConstants.Item2, "std");
var normalizedLayer = CNTKLib.PerDimMeanVarianceNormalize(inputLayer, mean, std);
var shape = new int[] { hiddenDim, 4 };
var weightParam = new Parameter(shape, DataType.Float, glorotInit, device, "wh");
var biasParam = new Parameter(new NDShape(1, hiddenDim), 0, device, "bh");
var hidLay = CNTKLib.Times(weightParam, normalizedLayer) + biasParam;
var hidLayerAct = CNTKLib.ReLU(hidLay);
var shapeOut = new int[] { 3, hiddenDim };
var wParamOut = new Parameter(shapeOut, DataType.Float, glorotInit, device, "wo");
var bParamOut = new Parameter(new NDShape(1, 3), 0, device, "bo");
var outLay = CNTKLib.Times(wParamOut, hidLayerAct) + bParamOut;
return outLay;
}
Complete Source Code Example
The whole source code about this example is listed below. The example shows how to normalize input feature for Iris famous data set. Notice that when using such way of data normalization, we don’t need to handle normalization for validation or testing data sets, because data normalization is part of the network model.
using System;
using System.Collections.Generic;
using System.Linq;
using System.Text;
using System.Threading.Tasks;
using CNTK;
namespace NormalizationLayerDemo
{
class Program
{
static string trainingDataPath = "./data/iris_training.txt";
static string validationDataPath = "./data/iris_validation.txt";
static void Main(string[] args)
{
DeviceDescriptor device = DeviceDescriptor.UseDefaultDevice();
var streamConfig = new StreamConfiguration[]
{
new StreamConfiguration("feature", 4),
new StreamConfiguration("flower", 3)
};
var feature = Variable.InputVariable(new NDShape(1, 4), DataType.Float, "feature");
var label = Variable.InputVariable(new NDShape(1, 3), DataType.Float, "flower");
var d = new Dictionary<StreamInformation, Tuple>();
using (var mbs = MinibatchSource.TextFormatMinibatchSource(
trainingDataPath , streamConfig, MinibatchSource.FullDataSweep,false))
{
d.Add(mbs.StreamInfo("feature"), new Tuple(null, null));
MinibatchSource.ComputeInputPerDimMeansAndInvStdDevs(mbs, d, device);
}
var ffnn_model = createFFModelWithNormalizationLayer
(feature,5,3,d.ElementAt(0).Value, device);
var trainingLoss = CNTKLib.CrossEntropyWithSoftmax
(new Variable(ffnn_model), label, "lossFunction");
var classError = CNTKLib.ClassificationError
(new Variable(ffnn_model), label, "classificationError");
var learningRatePerSample = new TrainingParameterScheduleDouble(0.01, 1);
var ll = Learner.SGDLearner(ffnn_model.Parameters(), learningRatePerSample);
var trainer = Trainer.CreateTrainer
(ffnn_model, trainingLoss, classError, new Learner[] { ll });
var mbsTraining = MinibatchSource.TextFormatMinibatchSource
(trainingDataPath, streamConfig, MinibatchSource.InfinitelyRepeat, true);
int epoch = 1;
while (epoch < 20)
{
var minibatchData = mbsTraining.GetNextMinibatch(65, device);
var arguments = new Dictionary
{
{ feature, minibatchData[mbsTraining.StreamInfo("feature")] },
{ label, minibatchData[mbsTraining.StreamInfo("flower")] }
};
trainer.TrainMinibatch(arguments, device);
if (minibatchData.Values.Any(a => a.sweepEnd))
{
reportTrainingProgress(feature, label, streamConfig, trainer, epoch, device);
epoch++;
}
}
Console.Read();
}
private static void reportTrainingProgress(Variable feature, Variable label,
StreamConfiguration[] streamConfig, Trainer trainer, int epoch, DeviceDescriptor device)
{
var mbsTrain = MinibatchSource.TextFormatMinibatchSource
(trainingDataPath, streamConfig, MinibatchSource.FullDataSweep, false);
var trainD = mbsTrain.GetNextMinibatch(int.MaxValue, device);
var a1 = new UnorderedMapVariableMinibatchData();
a1.Add(feature, trainD[mbsTrain.StreamInfo("feature")]);
a1.Add(label, trainD[mbsTrain.StreamInfo("flower")]);
var trainEvaluation = trainer.TestMinibatch(a1);
var mbsVal = MinibatchSource.TextFormatMinibatchSource
(validationDataPath, streamConfig, MinibatchSource.FullDataSweep, false);
var valD = mbsVal.GetNextMinibatch(int.MaxValue, device);
var a2 = new UnorderedMapVariableMinibatchData();
a2.Add(feature, valD[mbsVal.StreamInfo("feature")]);
a2.Add(label, valD[mbsVal.StreamInfo("flower")]);
var valEvaluation = trainer.TestMinibatch(a2);
Console.WriteLine($"Epoch={epoch},
Train Error={trainEvaluation}, Validation Error={valEvaluation}");
}
private static Function createFFModelWithNormalizationLayer
(Variable feature, int hiddenDim,int outputDim, Tuple avgStdConstants, DeviceDescriptor device)
{
var glorotInit = CNTKLib.GlorotUniformInitializer(
CNTKLib.DefaultParamInitScale,
CNTKLib.SentinelValueForInferParamInitRank,
CNTKLib.SentinelValueForInferParamInitRank, 1);
var inputLayer = feature;
var mean = new Constant(avgStdConstants.Item1, "mean");
var std = new Constant(avgStdConstants.Item2, "std");
var normalizedLayer = CNTKLib.PerDimMeanVarianceNormalize(inputLayer, mean, std);
var shape = new int[] { hiddenDim, 4 };
var weightParam = new Parameter(shape, DataType.Float, glorotInit, device, "wh");
var biasParam = new Parameter(new NDShape(1, hiddenDim), 0, device, "bh");
var hidLay = CNTKLib.Times(weightParam, normalizedLayer) + biasParam;
var hidLayerAct = CNTKLib.ReLU(hidLay);
var shapeOut = new int[] { 3, hiddenDim };
var wParamOut = new Parameter(shapeOut, DataType.Float, glorotInit, device, "wo");
var bParamOut = new Parameter(new NDShape(1, 3), 0, device, "bo");
var outLay = CNTKLib.Times(wParamOut, hidLayerAct) + bParamOut;
return outLay;
}
}
}
The output window should look like:

The data set files used in the example can be downloaded from here.
