Introduction
Good news everyone! You don't have to be a professor and you don't need a laboratory to make interesting experiments with Darwinian evolution. You only need to download this advanced cellular automaton to experience the raw power of natural selection first-hand.
Although there are many simulations which use genetic algorithms, I wanted to make a model that doesn't operate with arbitrary fitness functions and doesn't try to approach a pre-defined target outcome, but instead - just like real evolution in the living world - relies merely on successful reproductions. It was also important to make a model that is complex enough to make it possible to study different aspects of evolution and let organisms with different complexity compete against each other. At the same time, it had to be computation efficient to allow the user to simulate millions of organisms and hundreds of thousands of generations over a reasonable time frame. The good news is I think I've managed to create such a model.
The Model
Overview
The main actors of this simulation are the pixel-organisms. These are programmable creatures, that just like real organisms: compete for resources, consume what they can, digest (sort of), sense their environment, move, reproduce and when their time comes, eventually die. More importantly, they also evolve!
All of the listed activities and their surviving strategy/behavior is determined purely by their genes and as such also subject to mutation.
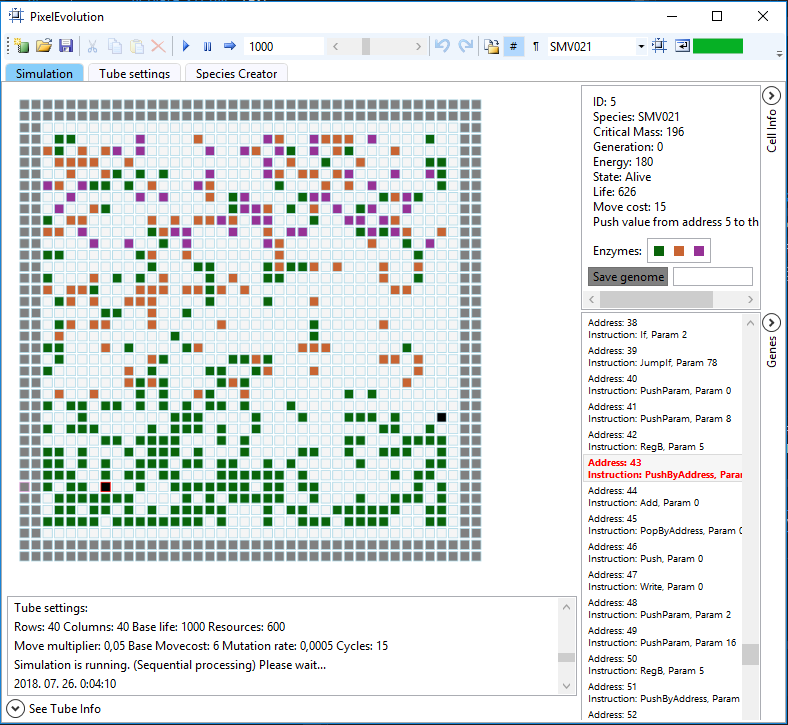
Figure 1: The pixel organism from the species SMV021 is represented by a black rectangle on the simulation field
The resources are represented as colored rectangles. There are 8000 possible types of resources, and each organism is only allowed to consume any of these resources if it has a corresponding digesting enzyme. (The details of this mechanism are elaborated in the architecture section.)
Natural Selection
The simulation field (aka test tube) is a closed eco-system, unless the user intervenes. Here, organisms are competing against each-other for the finite amount of resources, and when they consume enough, they are essentially converting resources to offspring. On the other hand, when an organism dies, it is converted back into chunks of resources. This is the cycle of life in the test tube. It's easy to see that those individuals who can't gather enough resources to leave offspring are weeded out by natural selection and their DNA is lost in history. (Or more precisely swiped by the garbage collector).
Random Mutation
As I mentioned, everything that a pixel-organism does is determined by its genes. Even movement, so a detrimental point mutation can cause a creature to stand in one spot waiting for its inevitable doom, or another one can cause it to run directly into the wall of the tube (which also means death). 3 types of mutations are used: point mutation, gene duplication/deletion and genome duplication.
Selective Pressures
There are many ways to create or change selective pressures for our pixel creatures. The most obvious one is to tweak the settings of the Tube:

Figure 2: Tube settings
For example, we can pressure the need for more complex decisions by increasing the cost of movement. Movement is a costly activity, in our model, it costs more processing cycle, and since each organism has equal processing cycles in every iteration (can be set by the "Cycles" property as seen in figure 2), they have to use it as efficiently as possible. In real life, it also costs more energy to actually move around in place than just looking around. If movement costs way more than sensing the environment or analyzing the sensory input, then our creatures better "think" before they move.
You can also see in figure 2 that there are 3 different regions defined. These regions are meant to represent differences between different local climates. By setting different types of resources for these regions, it's possible to create localized eco-systems (different selective pressures) with multiple species living in the same tube. It's also possible to see different species live in symbiosis if they are specialized in digesting completely different resources.
Genetic Drift
Genetic Drift can be emulated by choosing specific individuals from a population, then using the "save genome" feature (as seen in figure 1), then you can spawn instances of these in a new tube.
Architecture - The Anatomy of a Pixel-organism
One of the greatest challenges in designing this simulation was to create an architecture (virtual processor + machine code) which can handle mutable machine code. The traditional Van Neumann architecture is not suitable for this challenge. Random duplication or deletion of an instruction in a compiled machine code almost inevitably leads to unstable program and various exceptions. This is mainly because branching and jumping instructions point to defined memory addresses and duplication or deletion can shift all the values on those addresses, thus collapsing the branching structure of the program. For this reason, I designed a Gene based architecture.
Genes, Enzymes and Resources
Genes are essentially byte arrays with 3 elements. These elements are referred as codons. These genes are multi purpose entities: they can code for a digesting enzyme, code for instruction, or store data values. Their purpose is dependent on the context. In order to understand digesting enzymes, it's good to know that resources are also encoded by 3 byte codons. Each codon represents one of the RGB color channels, although the possible values are restricted. If each codon could get any value from 0 to 255, there would be more than 16 million possible outcomes 256*256*256. My goal was to make it feasible for an organism to randomly discover the enzyme it needs in a given environment and randomly mutating into a usable enzyme in 16 million possible options seemed a bit hopeless, so I reduced the problem space to 25*25*25 (25 range for each codon) 15625 options. (For the visualization purposes on the GUI, each value is multiplied by 10, to present nice, easily distinguishable colors). The basic structure of a Gene is illustrated in figure 3.

Figure 3: Structure of a gene

Figure 4: A resource selected on the GUI
The next problem considering the enzymes was that the first codon in a gene in an organism's genome is dedicated for addressing, so the 3 codons of a resource (Red codon, Green codon and Blue codon) had to be encoded by only 2 codons in a genomic gene. How is that possible? Thanks to the previously mentioned value restriction, it is possible. 2 bytes can represent 65536 different values which is more than enough to encode the 15625 possible resources. First, the 2 codons are interpreted as a base 125 number, then it is converted to a base 25 number thus we get the 3 resource codons we need.
When a new organism is born, its genotype is converted to a phenotype. This means (among other things) that all the genes that can be interpreted as a digesting enzymes is converted to base 25 codons - as described previously - and are added to an enzyme table. When this given organism tries to consume a resource, this enzyme table is checked for the corresponding values, and if there is a match, the mass of the resource is added to the energy of our creature and the resource disappears from the simulation field (it is consumed).
The Processor
The other thing that happens when the phenotype is created is that the genes are compiled to "machine code". In the machine code, the address byte becomes simply the index in the array holding the instructions and data, so a 2 dimensional byte array would be enough to store all the values we need from the genes. However, if we take a look at the Processor class, we see that the phenotype is a 3 dimensional jagged array. The reason behind this is that there is a problem with using a byte type for addressing instructions, namely that this way, no program could be more than 256 instructions long. To solve this issue, I had to implement a paging mechanism. The simple solution I've come up with was that all subroutines and methods are placed on dedicated different arrays so they each use separate addressing space. These separate arrays are called chromosomes in the model. Maybe it's worth noting that every concept so far introduced (including chromosomes) was based on its biological counterpart, except the processor. The closest thing would be comparing it to ribosomes since it also processes genes but it's still far from being accurate so let's stick with the term processor.
This virtual processor works very similar to real processors (the very primitive ones at least), It has a stack, a stack pointer, a program pointer, 3 registers (Reg A, Reg B, Reg C), the usual operations (excluding multiplication and division): Add, Subtract, Compare, Call, Push to stack, Pop from stack. Besides these, there are 3 simulation specific operations: Move(), Sense() and Consume(). I'm not planning to get into the nitty-gritty details of every aspect of the implementation, but it's important to share some information about these simulation specific functions, because they come in handy if you want to use the most interesting feature of the simulation: the species creator. (Figure 6)
Move
Movement is based on relative position to the organism. Basically, we are telling them to move up or down on the Y axis and right or left on the X axis. Intuitively movement to the left should be issued by negative values, and as usually computer graphics work, upwards on the Y axis (Up) would also be a negative value. In our model however, the code uses only unsigned byte values, so I had to make another compromise. As figure 5 shows (-)1 is coded by 101. When the Move() operation is called, the value in Reg A is loaded as a parameter for the X axis component of the movement, and the value of Reg C is loaded as the Y axis part (illustrated in figure 5 as well).

Figure 5: The selected organism will move one step upwards when the Move() function is called.
Sense
Sense() works in exactly the same way as movement as far as positioning is concerned. It can be used to survey neighboring cells for their resource content. It inherently checks the enzyme table as well: if the resource can't be consumed, the value 0 is loaded to Reg A. if it can be consumed, the mass value of the resource is loaded to Reg A. This allows the organism to make an informed decision on which way to move.
Consume
The Consume() operation makes the organism try to "eat" the resource on the cell it is currently standing on. As of now, there is no punishment if Consume() is called on a cell that has no resource or has resource, that our organism can't eat for the lack of a specific enzyme, although it is a wasted cycle. It may be changed in the future, to encourage less greedy behavior.
Lexer, Parser and Compiler
At the beginning, I mentioned that organisms are programmable, and of course I didn't mean that you must use this arbitrary gene-based machine code to be able to do that. I've designed a low level programming language Pbyte which is easy to use and has the basic features you'll need to write programs for these organisms or as I referred to: create species! For this reason, I had to implement a lexer, a recursive decent parser(link) and a compiler so readable Pbye programs can be compiled to Genes. I'm the first to admit, that neither the processor or the compiler is optimized, but this is a feature not a bug! Seriously, the whole system is designed to be subject to mutation and this includes the processor itself. The registers, the stack, the program pointer are all stored in the phenotype array so mutations introduced in the genotype can affect how they work. I won't get into the details of the actual test results I got from simulating millions of generations, but I have to say that the level of optimization through natural selection I witnessed surpassed all my expectations. A very simple species I created was compiled originally to 134 genes, after ~45000 generations faster and more viable organisms evolved with using only 55 genes.
Using the Application
I've tried my best to create a simple and efficient UI and provide tooltips when necessary, so figuring out the main functions shouldn't be an issue. To run a simulation, all you have to do is to select a cell on the simulation field, then hit F2 (this inserts a default organism into the test tube), then hit F10 to start the simulation. You can run some interesting experiments by tweaking the tube settings and trying out different enzyme and population setups, but the main feature of this program is creating and testing your own code/your own species.

Figure 6: Species creator
If you copy the following code to the species creator textbox, and hit F5 (compile) you should get a new, viable species in the templates dropdown menu. This demo code serves as a tutorial to introduce you to the process of species creation. I suggest to first test for yourself how it behaves before you continue with this article.
STR40 var v, x, y
{
x:=rega
y:=regc
v:=1
while(true)
{
if(y<3)
{
v:=1
}
if(y>37)
{
v:=101
}
rega:=1
regc:=0
while(x<37)
{
x:=x+1
Move()
Consume()
}
if(v==1)
{
y:=y+1
}
else
{
y:=y-1
}
rega:=0
regc:=v
Move()
rega:=101
regc:=0
while(x>2)
{
x:=x-1
Move()
Consume()
}
if(v==1)
{
y:=y+1
}
else
{
y:=y-1
}
rega:=0
regc:=v
Move()
}
}
When it's done, we can discuss how this behavior is encoded:
STR40 var v, x, y
x:=rega
y:=regc
v:=1
The first part is the initialization, the first string is going to be the name of our species. The name string is completely arbitrary of course, I named it STR40 because this organism is designed to work in a 40 * 40 tube, and it moves in a straight line.
This is followed by the variables we are going to use. X, and y are the coordinates we have to keep track of the position of our organism, the v variable decides if it is going upwards or downwards. As mentioned before, all organisms are born with their actual coordinates loaded in reg a, and reg c, so we can pass these values to the x, y variables (as named registers are used for other purposes).
while(true)
{
Above we have our main loop, the life cycle, this will run until the organism dies.
if(y<3)
{
v:=1
}
if(y>37)
{
v:=101
}
Here we have our main loop, the life cycle, this will run until the organism dies.
rega:=1
regc:=0
Here we setup Reg A and Reg C for movement. These values represent one step to the right.
while(x<37
{
x:=x+1
Move()
Consume()
}
This is the inner loop that first adjusts the x coordinate then makes the organism move and eat what it finds in each step, until it is too close to the right edge of the tube.
if(v==1)
{
y:=y+1
}
else
{
y:=y-1
}
rega:=0
regc:=v
Move()
When the right side of the tube reached, we have to change lines on the y axis (because we don't want to just move in the same line back and forth), which is determined by the previously setup v variable. This code first adjusts the y coordinate, then makes one step upwards or one step downwards, depending on the value of v. Remember: we are still too close to the right side of the tube.
rega:=101
regc:=0
while(x>2)
{
x:=x-1
Move()
Consume()
}
Here, we prepare Reg A and Reg C for movement to the left. The inner loop that follows is very similar to the one above, except the result will be one step to the left in each iteration. because we setup the registers that way.
if(v==1)
{
y:=y+1
}
else
{
y:=y-1
}
rega:=0
regc:=v
Move()
When the left side is reached, we change lines once again, and then the main loop (life cycle) can start all over again.
The main reason I publishing this app on this platform is, that I hope some of the programmers here take up the challenge and make some more viable species than the ones I've shared. If you have managed to do that, I encourage you to share your code in the comment section. Then, let the species compete against each other, just like in actual biology!
Technical POI
The GUI is implemented using WPF technology and accordingly I used the MVVM design pattern along with it. For the text editor element, I used AvalonEdit-WPF-Text-Editor.
For parallel processing, I came up with a solution, that made the application approximately 10 times faster:
The solutions required the implementation of a lexer, a parser and compiler. The algorithm for the parser is a recursive decent parser.
History
- 1st September, 2018: Initial version
