There is an offset between when a COVID case is reported and an eventual death. Using Python, SKLearn and a Jupyter notebook, find the offset in days between the two that results in the best correlation. The resulting linear equation can then accurately predict the number of future deaths based on already reported cases.
Introduction
The COVID Tracking Project publishes a daily, curated data set of global COVID cases, hospitizations, deaths and other data points. Reporting in the US seems to focus on lagging indicators like cases reported and fatalities-to-date. So using the Tracking Project's data, let's see the correlation between cases and future fatalities. As you might expect, there is a reliable relationship between the two.
Using the code
Python and Jupyter notebooks are great tools for experimentation and visualization of this sort of thing. Pandas, numpy, matplotlib and SKLearn provide more than enough functionality for a straightforward analysis.
startDate = np.datetime64('2020-06-24')
country = 'USA'
raw_data = pd.read_csv('https://covid.ourworldindata.org/data/owid-covid-data.csv',
usecols=['date', 'iso_code', 'new_deaths', 'new_cases', 'new_deaths_smoothed',
'new_cases_smoothed'], parse_dates=['date'])
df = raw_data[(raw_data.iso_code == country) & (raw_data.date >= startDate) &
(~raw_data.new_deaths.isnull()) &
(~raw_data.new_cases.isnull()) &
(~raw_data.new_deaths_smoothed.isnull()) &
(~raw_data.new_cases_smoothed.isnull())]
df.info()
After some initial exploration of the dataset for the US, it is pretty clear that there is a linear relationship between cases and deaths. From there, it's relatively straightforward to offset the case data from the fatalities by several days and find the offset with the best r2.
Model = namedtuple('Model', 'linearRegression r2 offset data')
def BestFitModel(new_cases, new_deaths, max_offset) -> Model:
best = Model(None, 0.0, 0, None)
for offset in range(1, max_offset):
cases = new_cases[-0:-offset]
deaths = new_deaths[offset:]
model = LinearRegression().fit(cases, deaths)
predictions = model.predict(cases)
r2 = metrics.r2_score(deaths, predictions)
if (r2 > best.r2):
best = Model(model, r2, offset,
pd.DataFrame({'predicted': predictions, 'actual': deaths}))
return best
Results
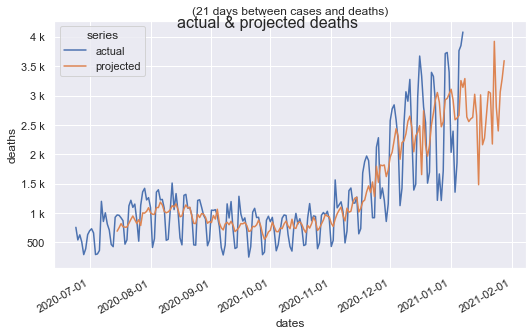
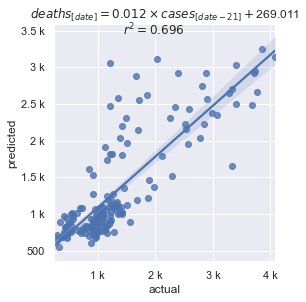
Reporting noise (weekends and holidays get under-reported) and are very spikey. Even with that noise, the data shows a strong correlation somewhere between 14 and 21 days offset. It's almost always a factor of 7 because of the weekend pattern.
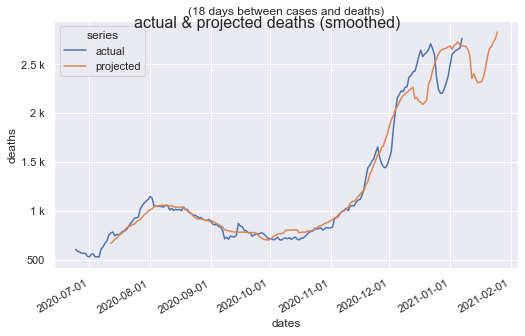
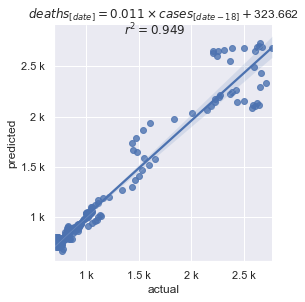
Smoothed data points, included in the Tracking Project's data set, removes the weekend pattern, but under-reporting over Thanksgiving and Christmas remain. Despite that, correlation is > 0.94.
Once the best fitting linear equation is found, it's straight forward to predict the number of deaths for the next number of days, equal to the offset used to find that equation.
def Predict(model: Model, dates, cases, deaths) -> pd.DataFrame:
minDate = np.amin(dates)
maxDate = np.amax(dates) + np.timedelta64(model.offset + 1, 'D')
projected_dates = [date for date in np.arange(minDate, maxDate, dt.timedelta(days=1))]
padding = pd.Series(np.full(model.offset, np.nan))
actual_deaths = deaths.append(padding)
projected_deaths = padding.append(pd.Series(model.linearRegression.predict(cases)))
frame = pd.DataFrame({"dates": projected_dates,
"actual": actual_deaths.values,
"projected": projected_deaths.values}, index=projected_dates)
return frame.melt(id_vars=['dates'], var_name='series', value_name='deaths')

Points of Interest
For the curious, the results are updated and published to GitHub each day.
History
- 2nd January, 2021 - Initial release
