Introduction
The systems biology is a subject that
studies the biology system and the behavior which comes from the interaction between
those components in this system. The major work in the systems biology study is
modeling the biological system and then using a program to simulate the model.
Because I am at hard working on the regulation signal transduction network
analysis in bacterial
Xanthomonas campestris pathovar carnpestris 8004 of my Microbial Genetics research job, and I need a
program to make the network simulation and data analysis. So I rewrite the
program PLAS which was written by Antonio
E.N.Fereira base on the research job of E.O.Voit for build up my own simulation system in the feature and
post my coding job here hopes it can helps other biological researchers.
Although PLAS have no LINUX edition
currently but the PLAS software works fine on LINUX platform using wine program.
PLAS software website: http://enzymology.fc.ul.pt/software/plas/
Background
Consider that A biochemical system has
N(N>=1) kinds substrate that involved in the reaction network, and the
temperature and volume of the system won’t be changed. So that we can using the
vector X(t) to represent that the state of this system in any time t:
X(t)=(x<sub>1</sub>(t), x<sub>2</sub>(t), ..., X<sub>N</sub>(t))
x<sub>i</sub>(t) represents the number or the concentration a
kind of substrate(i=1,2,…,N).
According to the view from the work of E.O.Voit,
a reaction procedure can be expressed as S-system function:
x<sub>i</sub><sup>'</sup>= α<sub>i</sub>∑(x<sub>j</sub>)<sup>gij</sup>-β<sub>i</sub>∑(x<sub>j</sub>)<sup>hij</sup>
x<sub>j</sub> represent the concentration of a kind of
substrate, parameters α<sub>i</sub> , <code>β<sub>i</sub> , g<sub>ij</sub> and <span lang="EN-US">h<sub>ij</sub></span><span lang="EN-US"> </span>are represent the that associated with the
reaction x<sub>i</sub>', they were all could achieved from
the laboratory experiment work, and the x<sub>i</sub>' is also represent the generation ratio of the
substrate x<sub>i</sub> .
So that a reaction system like
x<sub>1</sub> -> x<sub>2</sub> -> x<sub>3</sub>
Such a chain reaction can be express as
s-system function:
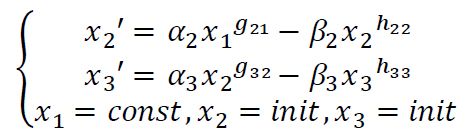
Because the left side of each equation x<sub>i</sub>' is represent as the substrate generation
ratio, so that after we calculate the value of the right side of the equation,
and then multiply it with the time unit, then we get the number or the
concentration change value of that substrate. And then we get the new
concentration of the substrate like the expression below:
x<sub>i</sub>=x<sub>i</sub>+x<sub>i</sub>'*△t
So that we could use the new concentration
to calculate other substrate in the next iteration loop.
All of this knowledge and more detail
information you can find out in the book <Computational Analysis of
Biochemical Systems> that E.O.Voit wrote or his scientific research articles
in the early year.
Using the code
Elements in the biochemical system can be
divided into 3 types: Substrate, reaction, and disturbing of the system.
A substrate you can treat it as a node in
the biochemical system network. And it gets two main attribute to stand for
this node: Name and concentration or amount value. The substrate class we
define in the model just for the storage of the calculation result.
Its data structure definition just simple
as you can see:
Public Class Var
<Xml.Serialization.XmlAttribute> Public Name As String
<Xml.Serialization.XmlAttribute> Public Value As Double
<Xml.Serialization.XmlAttribute> Public Title As String
<Xml.Serialization.XmlAttribute> Public Comment As String
Public Overrides Function ToString() As String
If String.IsNullOrEmpty(Comment) Then
Return String.Format("{0}={1}", IIf(Len(Title) > 0, Title, Name), Value)
Else
Return String.Format("{0}={1}; //{2}", IIf(Len(Title) > 0, Title, Name), Value, Comment)
End If
End Function
End Class
An equation stands for a biochemical
reaction, and it also means the interaction between the substrates (or the
system components). S-equation is a kind of chemical rate equation and uses it
for the concentration change rate of a substrate. So that the class equation
can basically define in two properties:
Public Class Equation
<Xml.Serialization.XmlAttribute> Public Name As String
<Xml.Serialization.XmlAttribute> Public Expression As String
End Class And it has a method to calculate the value of the equation expression:
Public Function Elapsed() As Boolean
Call Me.sBuilder.Clear()
sBuilder.Append(Expression)
For Each e In Kernel.Vars
Call sBuilder.Replace(e.Name, e.Value)
Next
Var.Value += (Microsoft.VisualBasic.Mathematical.Expression.Evaluate(sBuilder.ToString) * 0.1)
Return True
End Function
The
statement
Var.Value += (Microsoft.VisualBasic.Mathematical.Expression.Evaluate(sBuilder.ToString)
* 0.1)
in
this function is just do the iteration calculation: calculate the change rate
of the substrate using the equation expression an then multiply it with the
elapsed time to get the concentration change value, finally using the change
value to modify the current substrate concentration.
The disturbing of a biochemical system is
an important research method as it can let us get the important pathway or the
reaction in the system network. And also we can check a pathway or reaction is
exist or not through compare the calculation result or disturbing result in the
model with the result that come from the laboratory experiment.
Public Class Disturb
Public Enum Types
Increase
Decrease
ChangeTo
End Enum
<Xml.Serialization.XmlAttribute> Public Id As String
<Xml.Serialization.XmlAttribute> Public Start As Double
<Xml.Serialization.XmlAttribute> Public Interval As Double
<Xml.Serialization.XmlAttribute> Public Kicks As Integer
<Xml.Serialization.XmlAttribute> Public DisturbType As Types
<Xml.Serialization.XmlAttribute> Public Value As Double
End Class
What a disturb object have done to thewhole system in the system run time? The disturbing modifies the substrateconcentration directly on a specific time as the substrate concentration can representthe state of the whole system. And the disturbing have 3 type of modifymethods:
Public Shared ReadOnly Methods As System.Func(Of Double, Double, Double)() = {
Function(Var As Double, Delta As Double) Var + Delta,
Function(Var As Double, Delta As Double) Var - Delta,
Function(Var As Double, Delta As Double) Delta}
Public Enum Types
Increase
Decrease
ChangeTo
End Enum
The disturbing was managed by the Kicks
object, it gets two list objects, one is for the disturbing is not running and
another is for running disturbing object.
<Xml.Serialization.XmlIgnore> Friend PendingKicks As List(Of Disturb)
<Xml.Serialization.XmlIgnore> Friend RunningKicks As List(Of Disturb)
Script define format: Declaration syntax
are all write in the format like <Keyword> Expression, comment line are
those line first word is not in keyword list:
| Keyword
| Information
| Syntax
| Example
|
| RXN
| Declare a s-equation
| RXN <Substrate>=<Expression>
| RXN X1=10*X5-5*X1^0.5
|
| FUNC
| Declare a function
| FUNC <Name> <Expression>
| FUNC f x+y^2
|
| INIT
| Set the initial concentration value of a
substrate
| INIT <Substrate>=<value>
| INIT X1=2
|
| CONST
| Declare a constant
| CONST <Name> <value>
| CONST beta1 30
|
| NAMED
| Set the displaying title of a substrate,
optionally
| NAMED <Substrate> <Title>
| NAMED X1 ATP
|
| STIMULATE
| Set up a disturb in the system
| STIMULATE OBJECT <Substrate> START <Start
Time> KICKS <Kicks count> INTERVAL <Kicks Interval> VALUE <[Type]value>
| STIMULATE OBJECT X3 START 45 KICKS 30
INTERVAL 7 VALUE 5
|
| FINALTIME
| Set up the running time of the system
| FINALTIME <value>
| FINALTIME 10
|
| TITLE, COMMENT
| Model property define, optionally
| <Keyword> <VALUE>
| TITLE EXAMPLE SCRIPT
|
Here I define a Script class to compile the
script file to an available model to the kernel:

And here is an example script file:
RXN X1=10*X5*X3^-0.8-5*X1^0.5
RXN X2=5*X1^0.5-10*X2^0.5
RXN X3=2*X2^0.5-1.25*X3^0.5
RXN X4=8*X2^0.5-5*X4^0.5
INIT X1=2
INIT X2=0.25
INIT X3=0.64
INIT X4=0.64
INIT X5=0.5
STIMULATE OBJECT X1 START 4 KICKS 1 INTERVAL 1 VALUE 0
NAMED X1 ATP
NAMED X2 G6P
TITLE EXAMPLE SCRIPT
COMMENT NO COMMENT
FINALTIME 10
This script modeling such a branch and
feedback pathway:

The kernel class is tiny and easily to
extend, an array to hold the system state and an expression array to alter the
system state, a module to action the disturbing and a module to collecting the
output result, those 4 component were almost consist a complete kernel.
Public Class Kernel
Friend RtTime As Double
Friend Model As Model
Dim DataAcquisition As New DataAcquisition
Public Kicks As Kicks
Public Vars As Var()
Public Channels As Equation()
The kernel gets a method named Run to
action the biochemical system simulation: accumulate the system running time
from the start zero to the final time and the iteration calculation at each
loop. And yes, actually the state change of the whole system is the iteration
between each loop.
Public Sub Run()
For Me.RtTime = 0 To Model.FinalTime Step 0.1
Call Tick()
Next
End Sub
内核循环)
Public Sub Tick()
Dim Query As Generic.IEnumerable(Of Boolean) = From e As Equation In Channels Select e.Elapsed
Call DataAcquisition.Tick()
Call Kicks.Tick()
Query = Query.ToArray
End Sub
In fact, you also can get a parallel
calculation edition of this kernel just needs simply modify the LINQ query
statement as:
Dim Query As Generic.IEnumerable(Of Boolean) = From e As Equation In Channels.AsParallel Select e.Elapsed
But the parallel is not just always make the
things more efficient, sometimes it may slow down the system running because
the system needs to deal with more things. Or the parallel sometimes maybe make
the calculation data not so good.
The kernel gets a data acquisition class
object to collecting the simulation result and using the CSV file to store the data. And you can get the csv wrapper
class in the namespace <span lang="EN-US">Microsoft.VisualBasic.DataVisualization.Csv.</span><span lang="EN-US">File</span>
Public Class DataAcquisition
Dim _dataPackage As New Microsoft.VisualBasic.DataVisualization.Csv.File
Dim Kernel As Kernel
Public Sub Tick()
Dim Row As New Microsoft.VisualBasic.DataVisualization.Csv.File.Row
Call Row.Add(Kernel.RtTime)
For Each Var As Var In Kernel.Vars
Row.Add(Var.Value)
Next
_dataPackage.AppendLine(Row)
End Sub
Public Sub Save(Path As String)
Call _dataPackage.Save(Path)
End Sub
Public Sub [Set](Kernel As Kernel)
Dim Row As New Microsoft.VisualBasic.DataVisualization.Csv.File.Row
Me.Kernel = Kernel
Call Row.Add("Elapsed Time")
For Each Var As Var In Kernel.Vars
If Not String.IsNullOrEmpty(Var.Title) Then
Call Row.Add(String.Format("""{0}""", Var.Title))
Else
Call Row.Add(Var.Name)
End If
Next
Call _dataPackage.AppendLine(Row)
End Sub
Public Shared Function [Get](e As DataAcquisition) As Microsoft.VisualBasic.DataVisualization.Csv.File
Return e._dataPackage
End Function
End Class
Here I am using an example system to test
the program: Hull et al benchmarks / nonstiff systems.
Here is how the system defines:
RXN z1=2*(z1-z1*z2)
RXN z2=-1*(z2-z1*z2)
INIT z1=2
INIT z2=3
FINALTIME 20
Put this model into a text file, and then
using the code to run this system:
Sub Main()
Dim CSV = Kernel.Run(Script.Compile("./Hull.txt"))
CSV.Save("/home/xieguigang/Documents/Hull.csv")
End Sub
And then we open the CSV data file that we
get from the calculation, open in KingSoft Office or OpenOffice, using the data
to draw a scatter graph.

From the simulation result that we can see,
the concentration of two substrate z1 and z2 were change in period time, and it
get a nice period.
Points of Interest
The s-system was using a linear equation
collection to simulate a non-linear complexes system. Although the complexes
system was simplified by us, but we can still find some interesting attribute
through the simulation: The complexes system is sensitive to the initial value
of each component.
As we
can change the initial value of the component z1 in the hull system, and then
run the modified model to see what we get:

Z1=1

Z1=0

Z1=-1
From the change of the z1 initialize value
we can see that the system start to lose the ability of change the component
value in period time. And the most interesting thing is that when the z1=1, the
system has the attribute of period at the first time, but the thing goes crazy
at later: the continuing system calculation error accumulation finally change
the whole system behavior. And we also know this phenomenon its famous name:
The butterfly effect, the accumulated system error will finally change the
whole system behavior.
Although the
living system is also a complexes system, but the life system gets the ability
to restore itself from the disruption (We make the modifications of the system
initial value that is a disruption to the normal state), if the disruption is
not so deadly. The systems biology says that, the living system gets this
property: Emergent, Robustness, Complexity and Modularity. So from these points
of view, we could conclude that the biological living system gets a lot of
system error if we doing the calculation, and the biological system is not
sensitive to the precise values of biochemical parameters because of its
robustness, it can adjust itself from the error, but the mathematical equation do
not. Why the biological system not so sensitive, because it have found the way
to restore itself from the system error or to avoid the system crash, the
regulation network, which is really complexity. The regulation network is a
real complexes system and the mathematical equation is a fake one, that is the
reason why.
From this point of view that means the equations
is not enough to simulate a complex system. Although the mathematical equation
can gives the numeric result to us, but this numeric result just the
calculation result of a simplified complexes system.
So, based on the foundation work of E.O.Voit
and the concept & work mechanism of the S-system, we create a upgrade
version of the PLAS system, call it Object-S, not only the mathematical
equations in this system, and also the bio-macromolecule object, interaction
relationship and cell event integrated in this model. And I call this new
systems biology modeling project as the Genetic Clock Initiative project (GCI)
as the MICROSFT also have a bioinformatics research project for .NET named
Microsoft Biology Initiative (MBI) or Microsoft Biology Foundation (MBF).
This program was developing on Ubuntu
13.04(mono 2.1) and successfully debug and test on the Ubuntu LINUX and Windows
8 Enterprises.

