Hidden Markov Models (HMM) are stochastic methods to model temporal and sequence data. They are especially known for their application in temporal pattern recognition such as speech, handwriting, gesture recognition, part-of-speech tagging, musical score following, partial discharges and bioinformatics.
This code has also been incorporated in Accord.NET Framework, which includes the latest version of this code plus many other statistics and machine learning tools.
Contents
- Introduction
- Definition
- Notation
- Canonical problems
- Choosing the structure
- Algorithms
- Evaluation
- Decoding
- Learning
- Using the code
- Remarks
- Known issues
- Acknowledgements
- See also
- References
Hidden Markov Models were first described in a series of statistical papers by Leonard E. Baum and other authors in the second half of the 1960s. One of the first applications of HMMs was speech recognition, starting in the mid-1970s. Indeed, one of the most comprehensive explanations on the topic was published in “A Tutorial On Hidden Markov Models And Selected Applications in Speech Recognition”, by Lawrence R. Rabiner in 1989. In the second half of the 1980s, HMMs began to be applied to the analysis of biological sequences, in particular DNA. Since then, they have become ubiquitous in the field of bioinformatics.
Dynamical systems of discrete nature assumed to be governed by a Markov chain emit a sequence of observable outputs. Under the Markov assumption, it is also assumed that the latest output depends only on the current state of the system. Such states are often not known from the observer when only the output values are observable.
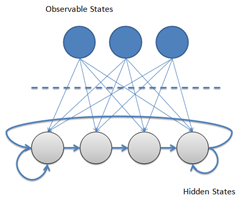
Hidden Markov Models attempt to model such systems and allow, among other things, (1) to infer the most likely sequence of states that produced a given output sequence, to (2) infer which will be the most likely next state (and thus predicting the next output) and (3) calculate the probability that a given sequence of outputs originated from the system (allowing the use of hidden Markov models for sequence classification).
The “hidden” in Hidden Markov Models comes from the fact that the observer does not know which state the system may be in, but has only a probabilistic insight on where it should be.
Hidden Markov Models can seem as finite state machines where for each sequence unit observation, there is a state transition and, for each state, there is an output symbol emission.
Traditionally, HMMs have been defined by the following quintuple:
)
where:
- N is the number of states for the model
- M is the number of distinct observations symbols per state, i.e., the discrete alphabet size
- A is the NxN state transition probability distribution given in the form of a matrix A = {aij}
- B is the NxM observation symbol probability distribution given in the form of a matrix B = {bj(k)}
- π is the initial state distribution vector π = {πi}
Note that if we opt out the structure parameters M and N, we have the more often used compact notation:
)
There are three canonical problems associated with hidden Markov models, which I'll quote from Wikipedia:
- Given the parameters of the model, compute the probability of a particular output sequence. This requires summation over all possible state sequences, but can be done efficiently using the Forward algorithm, which is a form of dynamic programming.
- Given the parameters of the model and a particular output sequence, find the state sequence that is most likely to have generated that output sequence. This requires finding a maximum over all possible state sequences, but can similarly be solved efficiently by the Viterbi algorithm.
- Given an output sequence or a set of such sequences, find the most likely set of state transition and output probabilities. In other words, derive the maximum likelihood estimate of the parameters of the HMM given a dataset of output sequences. No tractable algorithm is known for solving this problem exactly, but a local maximum likelihood can be derived efficiently using the Baum-Welch algorithm or the Baldi-Chauvin algorithm. The Baum-Welch algorithm is an example of a forward-backward algorithm, and is a special case of the Expectation-maximization algorithm.
The solution for those problems are exactly what makes Hidden Markov Models useful. The ability to learn from the data (using the solution of problem 3) and then become able to make predictions (solution to problem 2) and able to classify sequences (solution of problem 2) is nothing but applied machine learning. From this perspective, HMMs can just be seem as supervisioned sequence classifiers and sequence predictors with some other useful interesting properties.
Choosing the structure for a hidden Markov model is not always obvious. The number of states depend on the application and what interpretation one is willing to give to the hidden states. Some domain knowledge is required to build a suitable model and also to choose the initial parameters that an HMM can take. There is also some trial and error involved, and there are sometimes complex tradeoffs that have to be made between model complexity and difficulty of learning, just as is the case with most machine learning techniques.
Additional information can be found here.
The solution to the three canonical problems are the algorithms that make HMMs useful. Each of the three problems are described in the three subsections below.
The first canonical problem is the evaluation of the probability of a particular output sequence. It can be efficiently computed using either the Viterbi-forward or the Forward algorithms, both of which are forms of dynamic programming.
The Viterbi algorithm originally computes the most likely sequence of states which has originated a sequence of observations. In doing so, it is also able to return the probability of traversing this particular sequence of states. So to obtain Viterbi probabilities, please refer to the Decoding problem referred below.
The Forward algorithm, unlike the Viterbi algorithm, does not find a particular sequence of states; instead it computes the probability that any sequence of states has produced the sequence of observations. In both algorithms, a matrix is used to store computations about the possible state sequence paths that the model can assume. The forward algorithm also plays a key role in the Learning problem, and is thus implemented as a separate method.
public double Evaluate(int[] observations, bool logarithm)
{
if (observations == null)
throw new ArgumentNullException("observations");
if (observations.Length == 0)
return 0.0;
double likelihood = 0;
double[] coefficients;
forward(observations, out coefficients);
for (int i = 0; i < coefficients.Length; i++)
likelihood += Math.Log(coefficients[i]);
return logarithm ? likelihood : Math.Exp(likelihood);
}
private double[,] forward(int[] observations, out double[] c)
{
int T = observations.Length;
double[] pi = Probabilities;
double[,] A = Transitions;
double[,] fwd = new double[T, States];
c = new double[T];
for (int i = 0; i < States; i++)
c[0] += fwd[0, i] = pi[i] * B[i, observations[0]];
if (c[0] != 0)
{
for (int i = 0; i < States; i++)
fwd[0, i] = fwd[0, i] / c[0];
}
for (int t = 1; t < T; t++)
{
for (int i = 0; i < States; i++)
{
double p = B[i, observations[t]];
double sum = 0.0;
for (int j = 0; j < States; j++)
sum += fwd[t - 1, j] * A[j, i];
fwd[t, i] = sum * p;
c[t] += fwd[t, i];
}
if (c[t] != 0)
{
for (int i = 0; i < States; i++)
fwd[t, i] = fwd[t, i] / c[t];
}
}
return fwd;
}
The second canonical problem is the discovery of the most likely sequence of states that generated a given output sequence. This can be computed efficiently using the Viterbi algorithm. A trackback is used to detect the maximum probability path travelled by the algorithm. The probability of travelling such sequence is also computed in the process.
public int[] Decode(int[] observations, out double probability)
{
int T = observations.Length;
int states = States;
int minState;
double minWeight;
double weight;
int[,] s = new int[states, T];
double[,] a = new double[states, T];
for (int i = 0; i < states; i++)
{
a[i, 0] = (-1.0 * System.Math.Log(pi[i])) - System.Math.Log(B[i, observations[0]]);
}
for (int t = 1; t < T; t++)
{
for (int j = 0; j < states; j++)
{
minState = 0;
minWeight = a[0, t - 1] - System.Math.Log(A[0, j]);
for (int i = 1; i < states; i++)
{
weight = a[i, t - 1] - System.Math.Log(A[i, j]);
if (weight < minWeight)
{
minState = i;
minWeight = weight;
}
}
a[j, t] = minWeight - System.Math.Log(B[j, observations[t]]);
s[j, t] = minState;
}
}
minState = 0;
minWeight = a[0, T - 1];
for (int i = 1; i < states; i++)
{
if (a[i, T - 1] < minWeight)
{
minState = i;
minWeight = a[i, T - 1];
}
}
int[] path = new int[T];
path[T - 1] = minState;
for (int t = T - 2; t >= 0; t--)
path[t] = s[path[t + 1], t + 1];
probability = System.Math.Exp(-minWeight);
return path;
}
The third and last problem is the problem of learning the most likely parameters that best models a system given a set of sequences originated from this system. Most implementations I've seem did not consider the problem of learning from a set of sequences, but only from a single sequence at a time. The algorithm below, however, is fully suitable to learn from a set of sequences and also uses scaling, which is another thing I have not seen in other implementations.
The source code follows the original algorithm by Rabiner (1989). There are, however, some known issues with the algorithms detailed in Rabiner's paper. More information about those issues is available in another section of this article entitled “Remarks”.
public double Learn(int[][] observations, int iterations, double tolerance)
{
if (iterations == 0 && tolerance == 0)
throw new ArgumentException("Iterations and limit cannot be both zero.");
int N = observations.Length;
int currentIteration = 1;
bool stop = false;
double[] pi = Probabilities;
double[,] A = Transitions;
double[][, ,] epsilon = new double[N][, ,];
double[][,] gamma = new double[N][,];
for (int i = 0; i < N; i++)
{
int T = observations[i].Length;
epsilon[i] = new double[T, States, States];
gamma[i] = new double[T, States];
}
double oldLikelihood = Double.MinValue;
double newLikelihood = 0;
do
{
for (int i = 0; i < N; i++)
{
var sequence = observations[i];
int T = sequence.Length;
double[] scaling;
double[,] fwd = forward(observations[i], out scaling);
double[,] bwd = backward(observations[i], scaling);
for (int t = 0; t < T; t++)
{
double s = 0;
for (int k = 0; k < States; k++)
s += gamma[i][t, k] = fwd[t, k] * bwd[t, k];
if (s != 0)
{
for (int k = 0; k < States; k++)
gamma[i][t, k] /= s;
}
}
for (int t = 0; t < T - 1; t++)
{
double s = 0;
for (int k = 0; k < States; k++)
for (int l = 0; l < States; l++)
s += epsilon[i][t, k, l] = fwd[t, k] * A[k, l] *
bwd[t + 1, l] * B[l, sequence[t + 1]];
if (s != 0)
{
for (int k = 0; k < States; k++)
for (int l = 0; l < States; l++)
epsilon[i][t, k, l] /= s;
}
}
for (int t = 0; t < scaling.Length; t++)
newLikelihood += Math.Log(scaling[t]);
}
newLikelihood /= observations.Length;
if (checkConvergence(oldLikelihood, newLikelihood,
currentIteration, iterations, tolerance))
{
stop = true;
}
else
{
currentIteration++;
oldLikelihood = newLikelihood;
newLikelihood = 0.0;
for (int k = 0; k < States; k++)
{
double sum = 0;
for (int i = 0; i < N; i++)
sum += gamma[i][0, k];
pi[k] = sum / N;
}
for (int i = 0; i < States; i++)
{
for (int j = 0; j < States; j++)
{
double den = 0, num = 0;
for (int k = 0; k < N; k++)
{
int T = observations[k].Length;
for (int l = 0; l < T - 1; l++)
num += epsilon[k][l, i, j];
for (int l = 0; l < T - 1; l++)
den += gamma[k][l, i];
}
A[i, j] = (den != 0) ? num / den : 0.0;
}
}
for (int i = 0; i < States; i++)
{
for (int j = 0; j < Symbols; j++)
{
double den = 0, num = 0;
for (int k = 0; k < N; k++)
{
int T = observations[k].Length;
for (int l = 0; l < T; l++)
{
if (observations[k][l] == j)
num += gamma[k][l, i];
}
for (int l = 0; l < T; l++)
den += gamma[k][l, i];
}
B[i, j] = (num == 0) ? 1e-10 : num / den;
}
}
}
} while (!stop);
return newLikelihood;
}
Let's suppose we have gathered some sequences from a system we wish to model. The sequences are expressed as a integer array such as:
int[][] sequences = new int[][]
{
new int[] { 0,1,1,1,1,1,1 },
new int[] { 0,1,1,1 },
new int[] { 0,1,1,1,1 },
new int[] { 0,1, },
new int[] { 0,1,1 },
};
For us, it can be obvious to see that the system is outputting sequences that always start with a zero and have one or more ones at the end. But let's try to fit a Hidden Markov Model to predict those sequences.
HiddenMarkovModel hmm = new HiddenMarkovModel(2, 2);
hmm.Learn(sequences, 0.01);
Once the model is trained, let's test to see if it recognizes some sequences:
double l1 = hmm.Evaluate(new int[] { 0, 1 });
double l2 = hmm.Evaluate(new int[] { 0, 1, 1, 1 });
double l3 = hmm.Evaluate(new int[] { 1, 1 });
double l4 = hmm.Evaluate(new int[] { 1, 0, 0, 0 });
Of course, the model performs well as this is a rather simple example. A more useful test case would consist of allowing for some errors in the input sequences in the hope that the model will become more tolerant to measurement errors.
int[][] sequences = new int[][]
{
new int[] { 0,1,1,1,1,0,1,1,1,1 },
new int[] { 0,1,1,1,0,1,1,1,1,1 },
new int[] { 0,1,1,1,1,1,1,1,1,1 },
new int[] { 0,1,1,1,1,1 },
new int[] { 0,1,1,1,1,1,1 },
new int[] { 0,1,1,1,1,1,1,1,1,1 },
new int[] { 0,1,1,1,1,1,1,1,1,1 },
};
HiddenMarkovModel hmm = new HiddenMarkovModel(2, 3);
hmm.Learn(sequences, 0.0001);
double l1 = hmm.Evaluate(new int[] { 0,1 });
double l2 = hmm.Evaluate(new int[] { 0,1,1,1 });
double l3 = hmm.Evaluate(new int[] { 1,1 });
double l4 = hmm.Evaluate(new int[] { 1,0,0,0 });
double l5 = hmm.Evaluate(new int[] { 0,1,0,1,1,1,1,1,1 });
double l6 = hmm.Evaluate(new int[] { 0,1,1,1,1,1,1,0,1 });
We can see that despite having a very low probability, the likelihood values for the sequences containing a simulated measurement error are greater than the likelihoods for the sequences which do not follow the sequence structure at all.
In a subsequent article, we will see that those low values for the likelihoods will not be a problem because HMMs are often used in sets to form sequence classifiers. When used in such configurations, what really matters is which HMM returns the highest probability among others in the set.
A practical issue in the use of Hidden Markov Models to model long sequences is the numerical scaling of conditional probabilities. The probability of observing a long sequence given most models is extremely small, and the use of these extremely small numbers in computations often leads to numerical instability, making application of HMMs to genome length sequences quite challenging.
There are two common approaches to dealing with small conditional probabilities. One approach is to rescale the conditional probabilities using carefully designed scaling factors, and the other approach is to work with the logarithms of the conditional probabilities. For more information on using logarithms, please see the work entitled “Numerically Stable Hidden Markov Model Implementation”, by Tobias P. Mann.
The code on this article is based on the Tutorial by Rabiner. There are, however, some problems with the scaling and other algorithms. An errata depicting all issues is available in the website “An Erratum for ‘A Tutorial on Hidden Markov Models and Selected Applications in Speech Recognition’” and is maintained by Ali Rahimi. I have not yet verified if the implementation presented here also suffers from the same mistakes explained there. This code has worked well under many situations, but I cannot guarantee its perfectness. Please use at your own risk.
Thanks to Guilherme C. Pedroso, for the help with the Baum-Welch generalization for multiple input sequences. He has also co-written a very interesting article using hidden Markov models for gesture recognition, entitled “Automatic Recognition of Finger Spelling for LIBRAS based on a Two-Layer Architecture” published in the 25th Symposium On Applied Computing (ACM SAC 2010).
-
L. R. Rabiner, "A Tutorial on Hidden Markov Models, and Selected Applications in Speech Recognition," Proc. IEEE, Vol. 77, No. 2, pp. 257--286, Feb. 1989.
-
Warakagoda, Narada D., “A Hybrid ANN-HMM ASR system with NN based adaptive preprocessing”, Norges Tekniske Høgskole, 2006.
-
A. Rahimim, “An erratum for: A tutorial on hidden Markov models and selected applications in speech recognition”, World Wide Web. Available in: http://xenia.media.mit.edu/rahimi/rabiner/rabiner-errata/rabiner-errata.html.
-
T. P. Mann, "Numerically stable hidden markov model implementation," World Wide Web. Available in: http://www.phrap.org/compbio/mbt599/hmm_scaling_revised.pdf
-
Wikipedia contributors, "Hidden Markov model," Wikipedia, The Free Encyclopedia, http://en.wikipedia.org/wiki/Hidden_Markov_model (accessed March, 2010).
