This article introduces what data clustering is, and what it is good for. It provides the simplest possible implementation of the popular k-means++ algorithm in both FORTRAN and C, and discusses a couple of example problems.
What it is
Data clustering, sometimes called cluster analysis, is the process of finding groups in data. Objects that are alike are put together; objects that are different are separated.
As a graphical example, if your data is this:
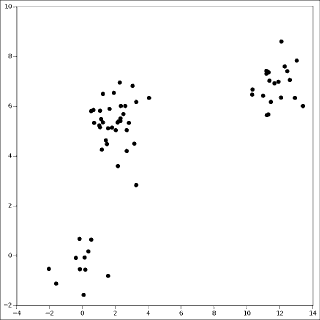
Then clustering is this:
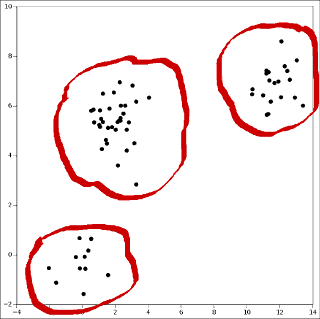
Data clustering is a topic in statistics and one of the major tasks of machine learning.
What it's good for
- Exploratory analysis:
- A company's marketing department may wish to place its customers in one of several market segments. A biologist may be interested in arranging a collection of specimens as several species. Much attention in statistics is given to hypothesis testing: to prove or disprove whether a difference exists between groups. Exploratory analysis is something else: there is no hypothesis yet. The analyst is finding what groups appear to exist.
- Summarize a data set:
- To cope with a large amount of data, often a small sample is taken to represent the full set. This sample may be chosen at random, but it is possible that rare objects will be overlooked. Data clustering can be used to produce a sample with both common and rare objects.
Data clustering should not be confused with classification. In classification, the classes are known beforehand. Usually a human expert provides a training set where each data point has been labeled with its class. In clustering, it is not known beforehand what classes might exist. For example, suppose that the data is information on some tumor cells. A classification task would be to identify each specimen as benign or malignant. A clustering task would be to identify distinct kinds of tumor cells.
Varieties of clustering
Prototype clustering
Each cluster is represented by a prototype, an example, or an average of some kind. Each object is compared to this prototype to decide if it belongs in the cluster. The k-means algorithm is of this variety.
Hierarchical clustering
Biologists place each species into a genus, each genus into a family, each family into an order, each order into a class, and each class into a division. Thus, all living things form a hierarchy of groups. Much early work in data clustering was done by botanists and was hierarchical, of course. It will not be discussed further here.
Graph clustering
Data points are compared to other points, and there are no cluster prototypes. Graph theory is invoked: the data objects are called vertices, and the similarities are called edges. Graph clustering is then a matter of cutting up the graph by slicing a minimum of edges. Since there is no central point, the clusters may be of any shape. This makes graph clustering useful for image segmentation, which will not be discussed further here.
Specifics
It becomes necessary specify exactly an "object" is, and what "similarity" means.
Let an object be a vector in RP, that is, an array of type float of length P. This is the easiest kind of data to work with, and many real problems of interest can be described this way.
x = [x1 x2…xP]
Let the prototype of each cluster be its centroid, the arithmetic mean of its vectors:
c = (1/N) Σxi
Let the dissimilarity between an object and its prototype be the Euclidean distance, that is, according to the Pythagorean Theorem:
d² = Σ(xj - cj)²
The k-means method
The k-means method has been around since 1956 or earlier, and is still widely used because it is simple, fast, and usually gives good results. Usually. Notice that formally, k-means is a method and not an algorithm, because it gives no guarantee that it will yield a correct answer.
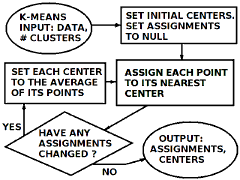
Call the objects "points" and the prototypes "centers." The method alternates between two main steps:
- Assign each point to its nearest center
- Let each center be the average of its points
Quit when the cluster assignments of the points quit changing. Let N denote the number of objects, P denote the measurements each object, and K denote the number of clusters sought. Then the greatest computation cost is in comparing the points to the centers, which requires O(NPK) operations each iteration.1
The k-means++ variant
The method as described above does not say how to choose the starting position. Traditional approaches have been to choose K points at random to be the initial centers, or to randomly assign each point to a cluster. It is possible for this to lead to very bad results, and the old advice was to run k-means several times and keep the best result.
Since 2006, new ways to choose the initial centers have been proposed that offer theoretical guarantees of approximating the optimal clustering.2, 3 This new variant is called the k-means++ algorithm. One point is chosen uniformly at random to be the first center. The remaining K-1 centers are chosen from the other points with a probability proportional to their squared distance to the nearest previously chosen center.
Here is how to choose an element with an arbitrary probability. First, we need a WORK array of length N to hold the shortest squared distance for each data point, and add up TOT as the sum of the elements in WORK. Then,4
CALL RANDOM_NUMBER (U)
U = U * TOT
TOT = 0.
DO I = 1, N
I1 = I
TOT = TOT + WORK(I)
IF (TOT > U) GO TO 20
END DO
20 DO J = 1, P
C(J,L) = X(J,I1)
END DO
Because every element in WORK is positive, adding turns an arbitrary probability distribution into a cumulative probability distribution, so that at each step of the loop on I, TOT must increase. As a graphic, it plots like this:
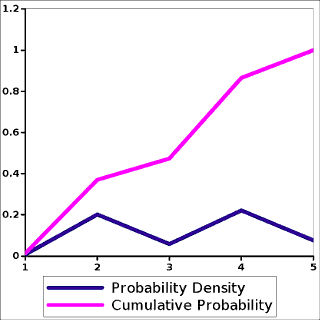
The cumulative distribution is the wand that can turn the mere uniform random number into a random variable of any other probability distribution.
Using the code
Subroutine KMPP implements k-means++ in plain, honest FORTRAN. A translation in C is provided. Here's the prototype:
SUBROUTINE KMPP (X, P, N, K, C, Z, WORK, IFAULT)
INTEGER P, N, K, Z, IFAULT
REAL X, C, WORK
DIMENSION X(P,N), C(P,K), Z(N), WORK(N)
where, X is the input data of points in RP, P is the number of variables, N is the number of objects, K is the number of clusters to be formed, C is the output centers as points in RP, Z is an array of integers from 1…K that denote the cluster membership of each point, WORK is used internally by the subroutine and its contents may be ignored by the user, and IFAULT is an error code that returns 0 on success.
In C, it looks like:
int kmpp (float **x, int p, int n, int k, float **c, int *z, float *work)
Notice that the matrices are in the form of arrays of pointers, that double precision is eschewed, and that the error code is now the return value of the function.
Example: the Iris data
The Iris data set is one of the oldest and best-known test problems in statistics.5 It was the example problem R. A. Fisher used to introduce Linear Discriminant Analysis in 1935. It is the measurements of the widths of petals, lengths of petals, widths of sepals, and lengths of sepals of 150 specimens of three species or iris. So N=150, P=4, K=3. The first two dimensions of the data plot as:
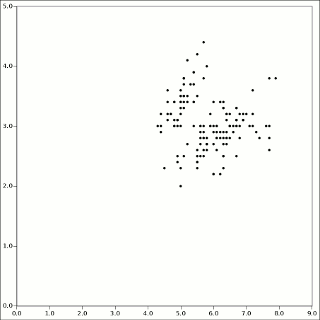
The t_iris example program calls KMPP and writes the results as a CSV file which is easy to import into any spreadsheet. With colors representing cluster membership and X marking the centers:
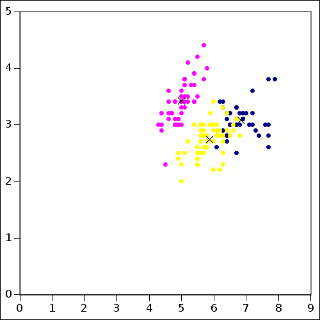
It is possible for the clusters to appear to overlap, because this plot is just a 2D projection of the 4-dimensional data set.6 For reference, here is the Iris data colored according to class label:
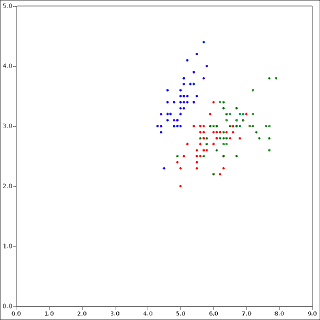
There are some disagreements. Remember, clustering is not classification. And k-means is pretty good, but it is certainly not perfect.
Example: random points in the unit circle
The t_round example program generates points at random in the unit circle, N=30000, P=2, K=6. The input to KMPP is:
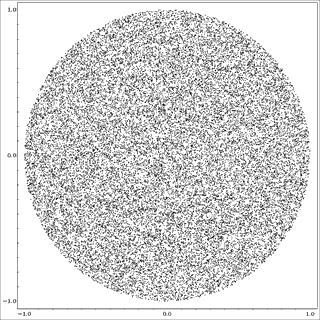
It is split up into clusters:
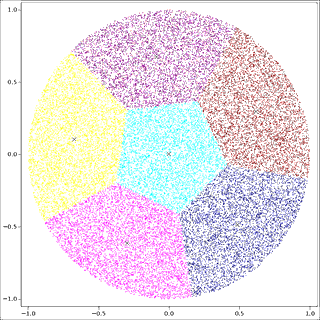
Notice that k-means obligingly partitions the data into K pieces, even if no clusters exist in the input!
Limitations
The simple routine given here
- Can't cope with data that is not represented as a vector of real numbers
- Can't cope with a data set from which any values are missing
- Can't tell you how many clusters you ought to make
- Can't measure the quality of its results
I hope in a future article to show at an Intermediate level of difficulty how these limitations may be addressed.
Notes
- If the centers are put into a tree data structure, then it is possible to find the nearest center to a point without comparing to all of them.
- Rafail Ostrovsky, Yuval Rabani, Leonard J. Schulman, and Chaitanya Swamy, "The Effectiveness of Lloyd-Type Methods for the k-means Problem", Proceedings of the 47th Annual IEEE Symposium on Foundations of Computer Science (FOCS'06)
- David Arthur and Sergei Vassilvitskii, "k-means++: the advantages of careful seeding", Proceedings of the eighteenth annual ACM-SIAM symposium on Discrete algorithms, 2007. Without doing any math, but using some common sense, it is obvious that because k-means++ depends on drawing random numbers, it can only "guarantee" the quality of the results in some average sense, over the long run. So it is still important to run the subroutine several times.
- This linear forward search could be replaced by storing the cumulative distribution in WORK, and doing a bisection search, for improved speed.
- The Iris data is not included in the distribution. You may download it from: The UC Irvine Machine Learning Repository, which has many valuable test problems.
- Singular Value Decomposition (SVD) or Multi-dimensional Scaling (MDS) can be used to find lower-dimension projections of a data set that are better than discarding entire dimensions.
References
- Leonard Kaufman and Peter J. Rousseeuw, Finding Groups in Data: An Introduction to Cluster Analysis, Wiley, 1990, reissued 2005.
- The R Core Team, kmeans.c, 2004.
- Wikipedia, of course
History
Version 1.0, 2 April 2017
