Introduction
As one of the oldest and easiest Machine Learning algorithms, implementing Simple Linear Regression can be an eye-opening and rewarding experience for anyone new to Machine Learning, Deep Learning and AI.
In this tutorial, we are going to implement Simple Linear Regression in Rust. To really internalize the algorithm, we won’t use any existing math or Machine Learning frameworks. The only external crate we will use is plotlib, which will allow us to visualize the results in a nice svg graphic.
You can find the source code for the entire project on https://github.com/phillikus/rust_ml.
Requirements
- Basic knowledge of the Rust programming language
- Basic high school math / statistic skills
Simple Linear Regression
The Linear Regression algorithm allows us to predict a dependent variable y based on a set of independent variables x0,x1..xn.
Based on a set of existing (x, y) pairs, our goal is to create a prediction function y(x):
y(x1..xn) = b0 + b1 * x1 + b2 * x2 + .. + bn * xn
where b0 is the so called intercept (y at x==0) and b1..bn are the coefficients that will be applied to the input values.
In the case of Simple Linear Regression, we have only one independent variable x, so we can simplify the above function to:
y(x) = b0 + b1 * x
Once we figure out the coefficient and the intercept, we can use them to make predictions for any new value of x by simply solving the equation. For example, if b0 == 5 and b1 == 2, y(4) can be calculated like this:
y(4) = 5 + 2 * 4 = 13
Example Data Set
To test our algorithm, I made up the following simple dataset:
[(1, 1), (2, 3), (3, 2), (4, 3), (5, 5)]
which can be visualized in the following scatter plot:
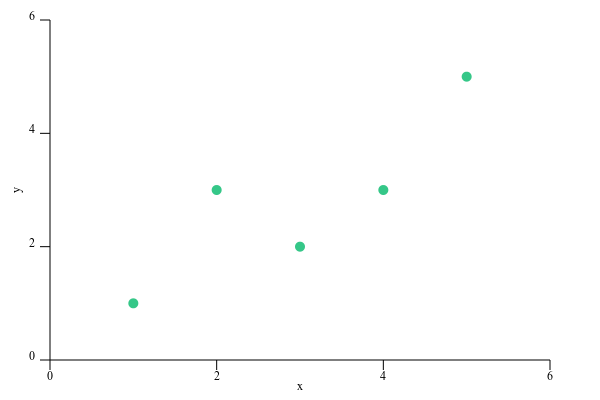
Our goal is to draw a straight line that is as close as possible to each of these points. Then, we can use that line to predict the y value for any x.
Estimating the Intercept and Coefficient
To estimate b0 and b1, we can use the following statistical equations:
b1 = Cov(x, y) / Var(x)
b0 = mean(y) - b1 * mean(x)
where Cov is the covariance, Var is the variance and mean is the mean of an array.
We can calculate all three of them with the following formulas:
mean(x) = sum(x) / length(x)
Var(x) = sum((x - mean(x))^2)
Cov = sum((x[i] - mean(x)) * (y[i] - mean(y)))
To learn more about the math behind this algorithm, take a look at Simple Linear Regression.
Project Structure
With all the basics layed out, let’s jump right into the code. I structured the project like this:

Our Linear Regression model will be implemented inside regression\linear_regression.rs. It will make use of utils\stat.rs, which contains statistic helper functions to calculate the mean, variance and covariance as defined above.
(The macros.rs module contains a helper function for unit tests, feel free to take a look at it in your own time).
To make our code usable as a library, we will reference the linear_regression module inside the lib.rs file and then use it as crate from main.rs.
Implementing Mean, Variance and Covariance
Let’s start by implementing the 3 math operations described above, as they make up the core of our algorithm. We will put them inside the stat.rs file:
pub fn mean(values : &Vec<f32>) -> f32 {
if values.len() == 0 {
return 0f32;
}
return values.iter().sum::<f32>() / (values.len() as f32);
}
pub fn variance(values : &Vec<f32>) -> f32 {
if values.len() == 0 {
return 0f32;
}
let mean = mean(values);
return values.iter()
.map(|x| f32::powf(x - mean, 2 as f32))
.sum::<f32>() / values.len() as f32;
}
pub fn covariance(x_values : &Vec<f32>, y_values : &Vec<f32>) -> f32 {
if x_values.len() != y_values.len() {
panic!("x_values and y_values must be of equal length.");
}
let length : usize = x_values.len();
if length == 0usize {
return 0f32;
}
let mut covariance : f32 = 0f32;
let mean_x = mean(x_values);
let mean_y = mean(y_values);
for i in 0..length {
covariance += (x_values[i] - mean_x) * (y_values[i] - mean_y)
}
return covariance / length as f32;
}
Note that we return 0 when an empty Vec<f32> is provided to either method. If size(x)!=size(y) in the covariance method, the code will break and panic. Beside that, the code should be pretty self-explanatory, it simply performs the calculations we defined above.
Implementing Simple Linear Regression
With our statistical functions ready, let’s dive into the actual algorithm. The interface of our Linear Regression model will look like this:
pub struct LinearRegression {
pub coefficient: Option<f32>,
pub intercept: Option<f32>
}
impl LinearRegression {
pub fn new() -> LinearRegression { .. }
pub fn fit(&mut self, x_values : &Vec<f32>, y_values : &Vec<f32>) { .. }
pub fn predict_list(&self, x_values : &Vec<f32>) -> Vec<f32> { .. }
pub fn predict(&self, x : f32) -> f32 { .. }
pub fn evaluate(&self, x_test : &Vec<f32>, y_test: &Vec<f32>) -> f32 { ..}
The struct contains two properties to get the intercept and coefficient of our model (b0 and b1). To initialize them, we will have to create an instance with the new function and then call its fit method (by default, both will be None).
Afterwards, we can use the other methods to make new predictions and evaluate the performance of our model (using Root Mean Squared Error).
Let’s go over these methods one by one:
pub fn new() -> LinearRegression {
LinearRegression { coefficient: None, intercept: None }
}
Nothing fancy going on here, we return a new instance of our struct with both the coefficient and intercept initialized to None.
Let’s dig into the fit function. Remember, that we can calculate b0 and b1 using statistics:
b1 = Cov(x, y) / Var(x)
b0 = mean(y) - b1 * mean(x)
Translated into Rust, our code will look like this:
pub fn fit(&mut self, x_values : &Vec<f32>, y_values : &Vec<f32>) {
let b1 = stat::covariance(x_values, y_values) / stat::variance(x_values);
let b0 = stat::mean(y_values) - b1 * stat::mean(x_values);
self.intercept = Some(b0);
self.coefficient = Some(b1);
}
All the heavy lifting is already implemented in stat.rs, so this code looks very straightforward. Remember to add use utils::stat; at the top of the file to make it compile.
To make predictions for new values of x, we can use the equation for Linear regression defined above:
y(x) = b0 + b1 * x
In Rust, this can be achieved easily:
pub fn predict(&self, x : f32) -> f32 {
if self.coefficient.is_none() || self.intercept.is_none() {
panic!("fit(..) must be called first");
}
let b0 = self.intercept.unwrap();
let b1 = self.coefficient.unwrap();
return b0 + b1 * x;
}
We first check if either the intercept (b0) and the coefficient (b1) is not initialized yet and display an error message in that case. Otherwise, we get b0 and b1 by unwrapping both properties and return the result of calculating y(x).
To make predictions for a list of inputs, I added an additional predict_list method:
pub fn predict_list(&self, x_values : &Vec<f32>) -> Vec<f32> {
let mut predictions = Vec::new();
for i in 0..x_values.len() {
predictions.push(self.predict(x_values[i]));
}
return predictions;
}
Here, we iterate over all input elements, predict their y-value and add it our list of predictions which will then be returned.
Performance Evaluation
All that is left now is the evaluate function, which will tell us how accurate our model is. Like mentioned above, we will use the Root Mean Squared Error method.
We can calculate the Root Mean Squared Error with the following formulas:
mse = sum((precition[i] - actual[i])^2)
rmse = sqrt(mse)
In Rust, this can be represented by the following function:
fn root_mean_squared_error(&self, actual : &Vec<f32>, predicted : &Vec<f32>) -> f32 {
let mut sum_error = 0f32;
let length = actual.len();
for i in 0..length {
sum_error += f32::powf(predicted[i] - actual[i], 2f32);
}
let mean_error = sum_error / length as f32;
return mean_error.sqrt();
}
Before we can use this function, however, we have to predict the values for our test set. We will do this inside the evaluate function, which then returns the result of the root_mean_squared_error function:
pub fn evaluate(&self, x_test : &Vec<f32>, y_test: &Vec<f32>) -> f32 {
if self.coefficient.is_none() || self.intercept.is_none() {
panic!("fit(..) must be called first");
}
let y_predicted = self.predict_list(x_test);
return self.root_mean_squared_error(y_test, &y_predicted);
}
Again, if either the coefficient or the intercept isn’t initialized, the code will break with an error message. Otherwise, we call the predict_list method to create a list of predictions and pass it to our rmse function.
Testing the Algorithm
With everything set up, it’s time to test our new Linear Regression algorithm! In the main.rs file, we can create a public main() function and initialize our model like this:
let mut model = linear_regression::LinearRegression::new();
let x_values = vec![1f32, 2f32, 3f32, 4f32, 5f32];
let y_values = vec![1f32, 3f32, 2f32, 3f32, 5f32];
model.fit(&x_values, &y_values);
We can use the methods we created to display the coefficient, intercept and accuracy:
println!("Coefficient: {0}", model.coefficient.unwrap());
println!("Intercept: {0}", model.intercept.unwrap());
println!("Accuracy: {0}", model.evaluate(&x_values, &y_values));
which will print:
Coefficient: 0.8
Intercept: 0.39999986
Accuracy: 0.69282037
To make predictions, we can use both our prediction methods:
let y_predictions : Vec<f32> = model.predict_list(&x_values);
let y_prediction : f32 = model.predict(4);
Visualizing Results
To see how well our algorithm performed without crunching numbers, it will be nice to get a visual representation of our predictions by plotting them into a 2D coordinate system. We can do so using the plotlib library:
let plot_actual = Scatter::from_vec(&actual)
.style(scatter::Style::new()
.colour("#35C788"));
let plot_prediction = Scatter::from_vec(&y_prediction)
.style(scatter::Style::new()
.marker(Marker::Square)
.colour("#DD3355"));
let v = View::new()
.add(&plot_actual)
.add(&plot_prediction)
.x_range(-0., 6.)
.y_range(0., 6.)
.x_label("x")
.y_label("y");
Page::single(&v).save("scatter.svg");
Don’t forget to import the crate and add them according to use directives:
extern crate plotlib;
use plotlib::scatter::Scatter;
use plotlib::scatter;
use plotlib::style::{Marker, Point};
use plotlib::view::View;
use plotlib::page::Page;
This will save a plot with the actual values and our predictions in a nicely formatted .svg file:

As you can see, our prediction matches the actual result quite well for most values. The only outliers are at x==2 and x==3, though both are still within acceptable bounds.
Conclusion
This concludes our trip to the Linear Regression algorithm, feel free to check out the source code and play around with different input values. Let me know if you have any questions, hints or comments.
