Introduction
I was interested in Needleman Wunsch Algorithm and I was searching for a good example in C#, but I had no luck. So I figured this might help someone.
Background
I wrote this based on this article.
There are three steps:
- Initialization step
- Matrix Fill step
- Traceback step
Let's say we have:
- sequence #1 GAATTCAGTTA and
- sequence #2 GGATCGA
- and scoring scheme is assumed
- Si,j = 2 if the residue at position i of sequence #1 is the same as the residue at position j of sequence #2 (match score); otherwise
- Si,j = -1 (mismatch score)
- w = -2 (gap penalty)
Initialization Step
The first row and the first column of the matrix can be initially filled with 0 as below:
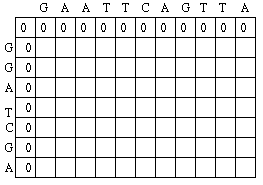
and the code would be as follows:
int refSeqCnt = refSeq.Length + 1;
int alineSeqCnt = alignSeq.Length + 1;
int[,] scoringMatrix = new int[alineSeqCnt, refSeqCnt];
for (int i = 0; i < alineSeqCnt; i++)
{
scoringMatrix[i, 0] = 0;
}
for (int j = 0; j < refSeqCnt; j++)
{
scoringMatrix[0, j] = 0;
}
Matrix Fill Step
For each position, Mi,j is defined to be the maximum score at position i,j.
Mi,j = Maximum[
Mi-1,j-1 + Si,j (Match/mismatch in the diagonal),
Mi,j-1 + w (gap in sequence #1)
Mi-1,j + w (gap in sequence #2)

for (int i = 1; i < alineSeqCnt; i++)
{
for (int j = 1; j < refSeqCnt; j++)
{
int scroeDiag = 0;
if (refSeq.Substring(j - 1, 1) == alignSeq.Substring(i - 1, 1))
scroeDiag = scoringMatrix[i - 1, j - 1] + 2;
else
scroeDiag = scoringMatrix[i - 1, j - 1] + -1;
int scroeLeft = scoringMatrix[i, j - 1] - 2;
int scroeUp = scoringMatrix[i - 1, j] - 2;
int maxScore = Math.Max(Math.Max(scroeDiag, scroeLeft), scroeUp);
scoringMatrix[i, j] = maxScore;
}
}
Traceback Step
After the matrix fill step, the maximum global alignment score for the two sequences is 3. The traceback will determine the actual alignment(s) that results in the maximum score.
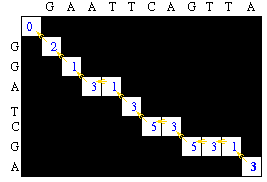
This gives alignment of:
char[] alineSeqArray = alignSeq.ToCharArray();
char[] refSeqArray = refSeq.ToCharArray();
string AlignmentA = string.Empty;
string AlignmentB = string.Empty;
int m = alineSeqCnt - 1;
int n = refSeqCnt - 1;
while (m > 0 && n > 0)
{
int scroeDiag = 0;
if (alineSeqArray[m - 1] == refSeqArray[n - 1])
scroeDiag = 2;
else
scroeDiag = -1;
if (m > 0 && n > 0 && scoringMatrix[m, n] == scoringMatrix[m - 1, n - 1] + scroeDiag)
{
AlignmentA = refSeqArray[n - 1] + AlignmentA;
AlignmentB = alineSeqArray[m - 1] + AlignmentB;
m = m - 1;
n = n - 1;
}
else if (n > 0 && scoringMatrix[m, n] == scoringMatrix[m, n - 1] - 2)
{
AlignmentA = refSeqArray[n - 1] + AlignmentA;
AlignmentB = "-" + AlignmentB;
n = n - 1;
}
else
{
AlignmentA = "-" + AlignmentA;
AlignmentB = alineSeqArray[m - 1] + AlignmentB;
m = m - 1;
}
}
I tested with 14739 bp sequence but it was a bit slow. If you have a better and faster solution, please let me know. I just wanted to share this with someone who might be interested in this algorithm in C#. You are welcome to give me any advice or suggestions.
Now I would like to research the multiple sequence alignment.
